diff --git a/.github/CONTRIBUTING.md b/.github/CONTRIBUTING.md
new file mode 100644
index 0000000..6f7efed
--- /dev/null
+++ b/.github/CONTRIBUTING.md
@@ -0,0 +1,262 @@
+# Contributing to XAVIER
+
+## Proposing changes with issues
+
+If you want to make a change, it's a good idea to first
+[open an issue](https://code-review.tidyverse.org/issues/)
+and make sure someone from the team agrees that it’s needed.
+
+If you've decided to work on an issue,
+[assign yourself to the issue](https://docs.github.com/en/issues/tracking-your-work-with-issues/assigning-issues-and-pull-requests-to-other-github-users#assigning-an-individual-issue-or-pull-request)
+so others will know you're working on it.
+
+## Pull request process
+
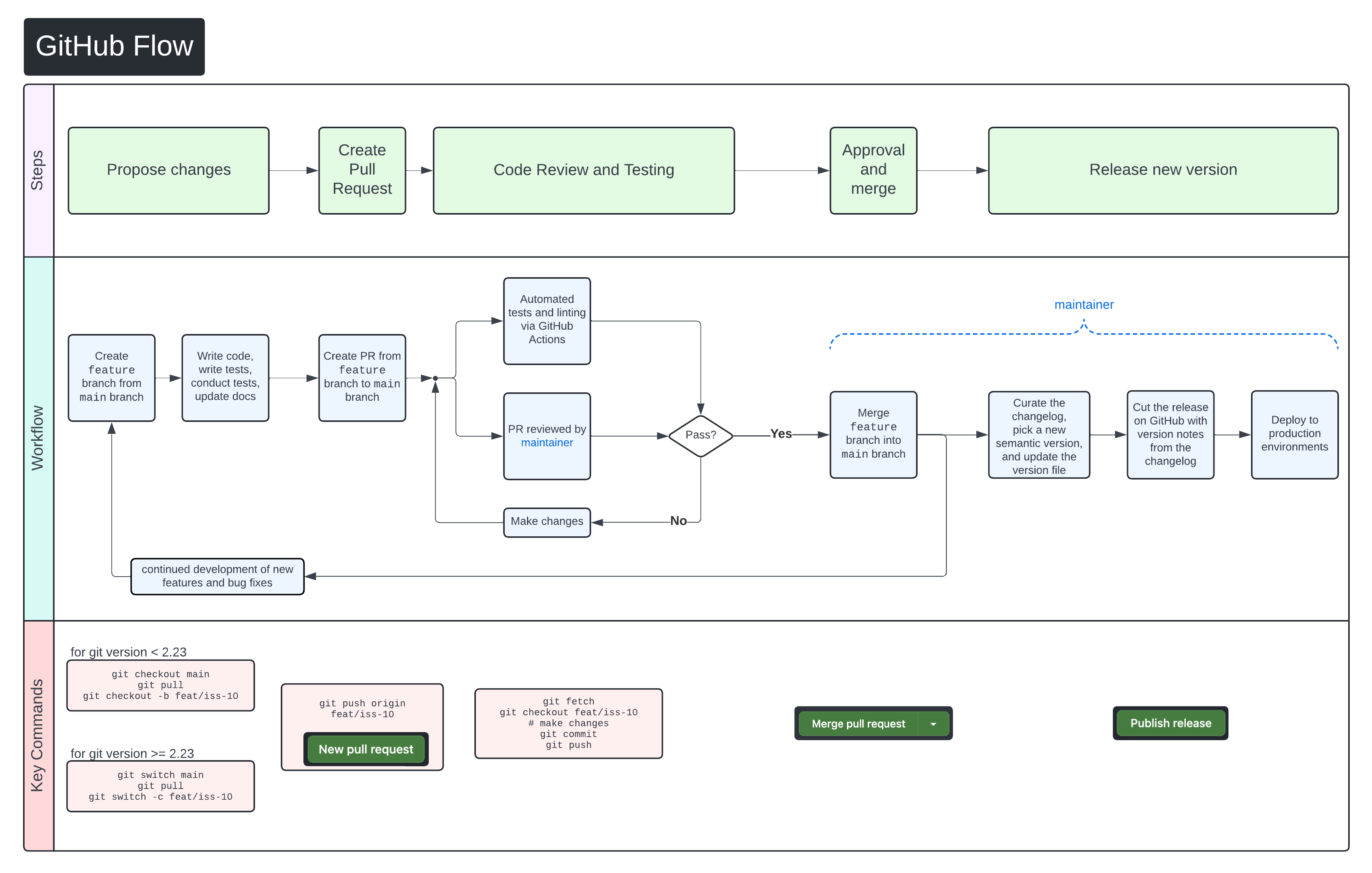
+We use [GitHub Flow](https://docs.github.com/en/get-started/using-github/github-flow)
+as our collaboration process.
+Follow the steps below for detailed instructions on contributing changes to
+XAVIER.
+
+
+
+### Clone the repo
+
+If you are a member of [CCBR](https://github.com/CCBR),
+you can clone this repository to your computer or development environment.
+Otherwise, you will first need to
+[fork](https://docs.github.com/en/pull-requests/collaborating-with-pull-requests/working-with-forks/fork-a-repo)
+the repo and clone your fork. You only need to do this step once.
+
+```sh
+git clone https://github.com/CCBR/XAVIER
+```
+
+> Cloning into 'XAVIER'...
+> remote: Enumerating objects: 1136, done.
+> remote: Counting objects: 100% (463/463), done.
+> remote: Compressing objects: 100% (357/357), done.
+> remote: Total 1136 (delta 149), reused 332 (delta 103), pack-reused 673
+> Receiving objects: 100% (1136/1136), 11.01 MiB | 9.76 MiB/s, done.
+> Resolving deltas: 100% (530/530), done.
+
+```sh
+cd XAVIER
+```
+
+### If this is your first time cloning the repo, you may need to install dependencies
+
+- Install snakemake and singularity or docker if needed (biowulf already has these available as modules).
+
+- Install the python dependencies with pip
+
+ ```sh
+ pip install .
+ ```
+
+ If you're developing on biowulf, you can use our shared conda environment which already has these dependencies installed
+
+ ```sh
+ . "/data/CCBR_Pipeliner/db/PipeDB/Conda/etc/profile.d/conda.sh"
+ conda activate py311
+ ```
+
+- Install [`pre-commit`](https://pre-commit.com/#install) if you don't already
+ have it. Then from the repo's root directory, run
+
+ ```sh
+ pre-commit install
+ ```
+
+ This will install the repo's pre-commit hooks.
+ You'll only need to do this step the first time you clone the repo.
+
+### Create a branch
+
+Create a Git branch for your pull request (PR). Give the branch a descriptive
+name for the changes you will make, such as `iss-10` if it is for a specific
+issue.
+
+```sh
+# create a new branch and switch to it
+git branch iss-10
+git switch iss-10
+```
+
+> Switched to a new branch 'iss-10'
+
+### Make your changes
+
+Edit the code, write and run tests, and update the documentation as needed.
+
+#### test
+
+Changes to the **python package** code will also need unit tests to demonstrate
+that the changes work as intended.
+We write unit tests with pytest and store them in the `tests/` subdirectory.
+Run the tests with `python -m pytest`.
+
+If you change the **workflow**, please run the workflow with the test profile
+and make sure your new feature or bug fix works as intended.
+
+#### document
+
+If you have added a new feature or changed the API of an existing feature,
+you will likely need to update the documentation in `docs/`.
+
+### Commit and push your changes
+
+If you're not sure how often you should commit or what your commits should
+consist of, we recommend following the "atomic commits" principle where each
+commit contains one new feature, fix, or task.
+Learn more about atomic commits here:
+
+
+First, add the files that you changed to the staging area:
+
+```sh
+git add path/to/changed/files/
+```
+
+Then make the commit.
+Your commit message should follow the
+[Conventional Commits](https://www.conventionalcommits.org/en/v1.0.0/)
+specification.
+Briefly, each commit should start with one of the approved types such as
+`feat`, `fix`, `docs`, etc. followed by a description of the commit.
+Take a look at the [Conventional Commits specification](https://www.conventionalcommits.org/en/v1.0.0/#summary)
+for more detailed information about how to write commit messages.
+
+```sh
+git commit -m 'feat: create function for awesome feature'
+```
+
+pre-commit will enforce that your commit message and the code changes are
+styled correctly and will attempt to make corrections if needed.
+
+> Check for added large files..............................................Passed
+> Fix End of Files.........................................................Passed
+> Trim Trailing Whitespace.................................................Failed
+>
+> - hook id: trailing-whitespace
+> - exit code: 1
+> - files were modified by this hook
>
+> Fixing path/to/changed/files/file.txt
>
+> codespell................................................................Passed
+> style-files..........................................(no files to check)Skipped
+> readme-rmd-rendered..................................(no files to check)Skipped
+> use-tidy-description.................................(no files to check)Skipped
+
+In the example above, one of the hooks modified a file in the proposed commit,
+so the pre-commit check failed. You can run `git diff` to see the changes that
+pre-commit made and `git status` to see which files were modified. To proceed
+with the commit, re-add the modified file(s) and re-run the commit command:
+
+```sh
+git add path/to/changed/files/file.txt
+git commit -m 'feat: create function for awesome feature'
+```
+
+This time, all the hooks either passed or were skipped
+(e.g. hooks that only run on R code will not run if no R files were
+committed).
+When the pre-commit check is successful, the usual commit success message
+will appear after the pre-commit messages showing that the commit was created.
+
+> Check for added large files..............................................Passed
+> Fix End of Files.........................................................Passed
+> Trim Trailing Whitespace.................................................Passed
+> codespell................................................................Passed
+> style-files..........................................(no files to check)Skipped
+> readme-rmd-rendered..................................(no files to check)Skipped
+> use-tidy-description.................................(no files to check)Skipped
+> Conventional Commit......................................................Passed
> [iss-10 9ff256e] feat: create function for awesome feature
+> 1 file changed, 22 insertions(+), 3 deletions(-)
+
+Finally, push your changes to GitHub:
+
+```sh
+git push
+```
+
+If this is the first time you are pushing this branch, you may have to
+explicitly set the upstream branch:
+
+```sh
+git push --set-upstream origin iss-10
+```
+
+> Enumerating objects: 7, done.
+> Counting objects: 100% (7/7), done.
+> Delta compression using up to 10 threads
+> Compressing objects: 100% (4/4), done.
+> Writing objects: 100% (4/4), 648 bytes | 648.00 KiB/s, done.
+> Total 4 (delta 3), reused 0 (delta 0), pack-reused 0
+> remote: Resolving deltas: 100% (3/3), completed with 3 local objects.
+> remote:
+> remote: Create a pull request for 'iss-10' on GitHub by visiting:
+> remote: https://github.com/CCBR/XAVIER/pull/new/iss-10
+> remote:
+> To https://github.com/CCBR/XAVIER
>
> [new branch] iss-10 -> iss-10
+> branch 'iss-10' set up to track 'origin/iss-10'.
+
+We recommend pushing your commits often so they will be backed up on GitHub.
+You can view the files in your branch on GitHub at
+`https://github.com/CCBR/XAVIER/tree/`
+(replace `` with the actual name of your branch).
+
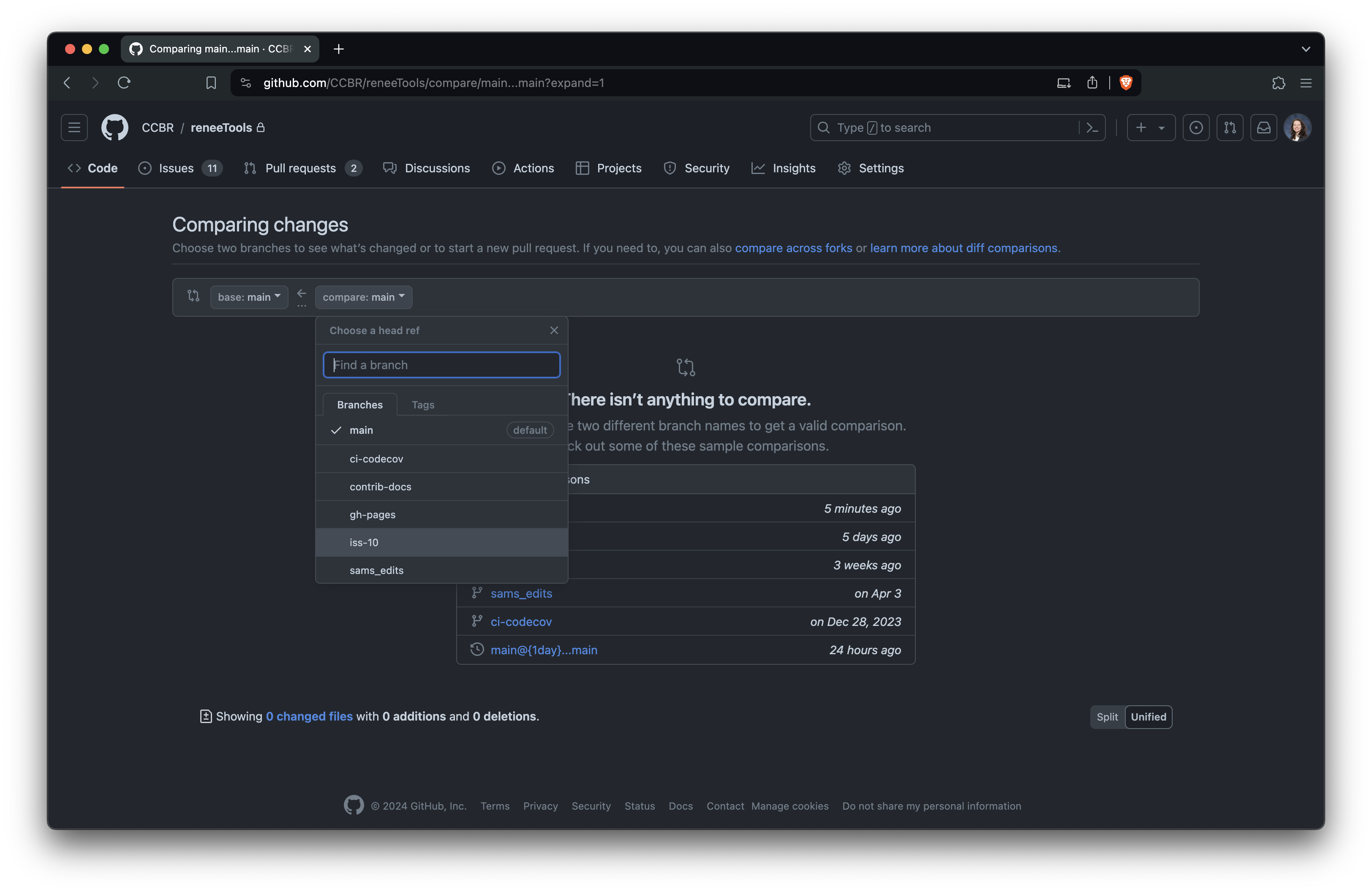
+### Create the PR
+
+Once your branch is ready, create a PR on GitHub:
+
+
+Select the branch you just pushed:
+
+
+
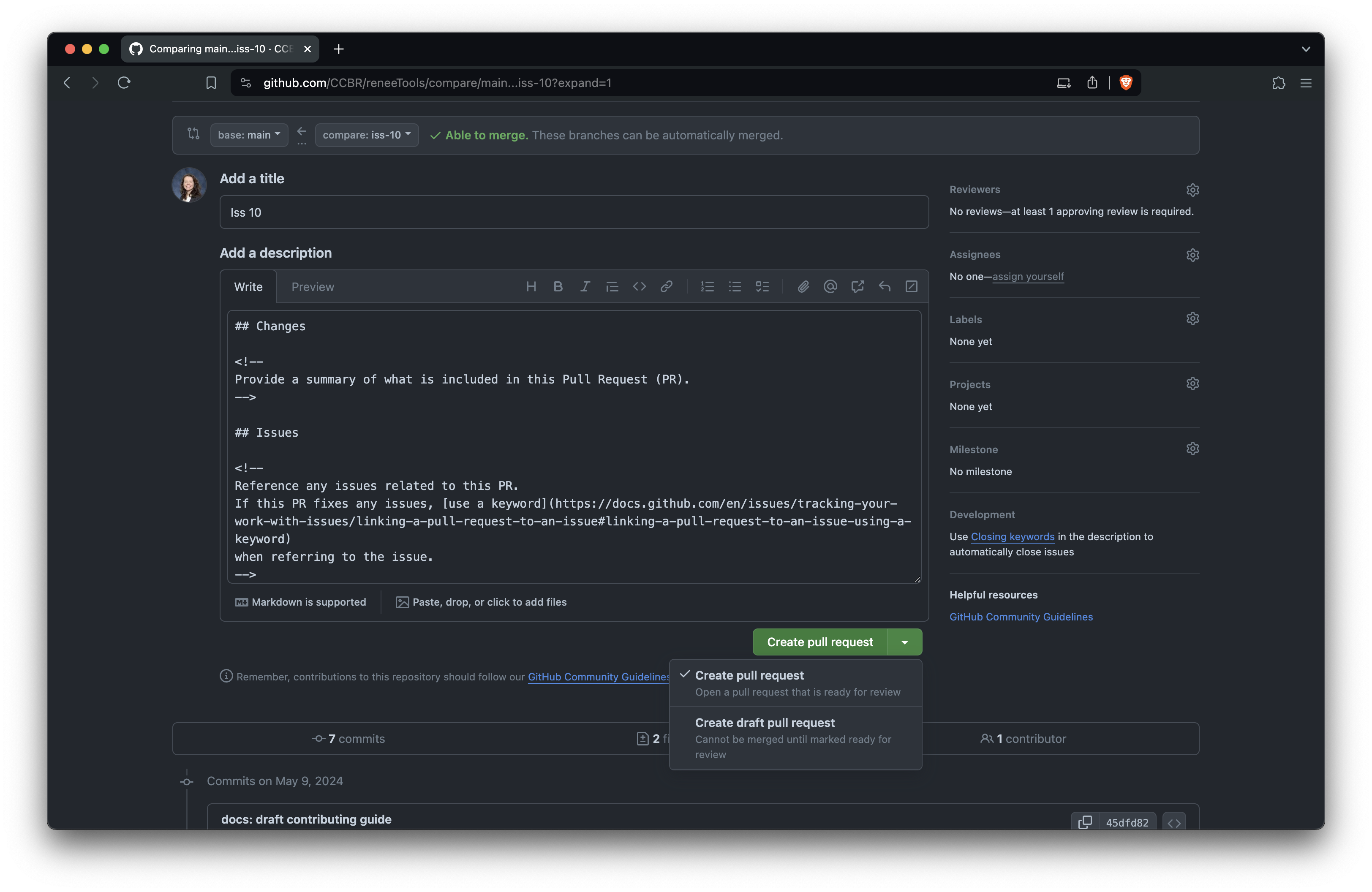
+Edit the PR title and description.
+The title should briefly describe the change.
+Follow the comments in the template to fill out the body of the PR, and
+you can delete the comments (everything between ``) as you go.
+Be sure to fill out the checklist, checking off items as you complete them or
+striking through any irrelevant items.
+When you're ready, click 'Create pull request' to open it.
+
+
+
+Optionally, you can mark the PR as a draft if you're not yet ready for it to
+be reviewed, then change it later when you're ready.
+
+### Wait for a maintainer to review your PR
+
+We will do our best to follow the tidyverse code review principles:
+.
+The reviewer may suggest that you make changes before accepting your PR in
+order to improve the code quality or style.
+If that's the case, continue to make changes in your branch and push them to
+GitHub, and they will appear in the PR.
+
+Once the PR is approved, the maintainer will merge it and the issue(s) the PR
+links will close automatically.
+Congratulations and thank you for your contribution!
+
+### After your PR has been merged
+
+After your PR has been merged, update your local clone of the repo by
+switching to the main branch and pulling the latest changes:
+
+```sh
+git checkout main
+git pull
+```
+
+It's a good idea to run `git pull` before creating a new branch so it will
+start from the most recent commits in main.
+
+## Helpful links for more information
+
+- [GitHub Flow](https://docs.github.com/en/get-started/using-github/github-flow)
+- [semantic versioning guidelines](https://semver.org/)
+- [changelog guidelines](https://keepachangelog.com/en/1.1.0/)
+- [tidyverse code review principles](https://code-review.tidyverse.org)
+- [reproducible examples](https://www.tidyverse.org/help/#reprex)
+- [nf-core extensions for VS Code](https://marketplace.visualstudio.com/items?itemName=nf-core.nf-core-extensionpack)
diff --git a/.github/workflows/build-docker-meta.yml b/.github/workflows/build-docker-meta.yml
new file mode 100644
index 0000000..87a61bc
--- /dev/null
+++ b/.github/workflows/build-docker-meta.yml
@@ -0,0 +1,46 @@
+name: build-docker-meta
+
+on:
+ workflow_dispatch:
+ inputs:
+ context_dir:
+ type: string
+ description: path to the directory containing the Dockerfile and meta.yml file (e.g. common/ccbr_chipseeker/)
+ required: true
+ push:
+ type: boolean
+ description: Push to DockerHub (uncheck to only build the container without pushing)
+ required: true
+ default: true
+
+jobs:
+ build-docker:
+ runs-on: ubuntu-latest
+ steps:
+ - uses: actions/checkout@v4
+ - uses: pietrobolcato/action-read-yaml@1.0.0
+ id: metadata
+ with:
+ config: ${{ github.workspace }}/${{ github.event.inputs.context_dir }}/meta.yml
+ - name: Login to DockerHub
+ if: ${{ github.event.inputs.push == 'true' }}
+ uses: docker/login-action@v2
+ with:
+ username: ${{ secrets.DOCKERHUB_USERNAME }}
+ password: ${{ secrets.DOCKERHUB_TOKEN }}
+ - name: Prepare build-time variables
+ id: vars
+ run: |
+ echo "DATE=$(date +"%Y-%m-%d")" >> "$GITHUB_OUTPUT"
+ - name: Build and push
+ uses: docker/build-push-action@v4
+ # only try building & pushing the container if parsing the metadata worked
+ if: ${{ steps.metadata.outputs['container'] != '' }}
+ with:
+ context: ${{ github.event.inputs.context_dir }}
+ push: ${{ github.event.inputs.push }}
+ tags: ${{ steps.metadata.outputs['container'] }}
+ build-args: |
+ BUILD_DATE=${{ steps.vars.outputs.DATE }}
+ BUILD_TAG=${{ steps.metadata.outputs['version'] }}
+ REPONAME=${{ steps.metadata.outputs['image_name'] }}
diff --git a/.github/workflows/docker.yml b/.github/workflows/docker.yml
new file mode 100644
index 0000000..de1d9f8
--- /dev/null
+++ b/.github/workflows/docker.yml
@@ -0,0 +1,64 @@
+name: docker
+
+on:
+ push:
+ branches:
+ - main
+ paths:
+ - "docker/**"
+ pull_request:
+ branches:
+ - main
+ paths:
+ - "docker/**"
+ workflow_dispatch:
+
+jobs:
+ generate-matrix:
+ runs-on: ubuntu-latest
+ outputs:
+ matrix-metadata: ${{ steps.metadata.outputs.matrix }}
+ steps:
+ - uses: hellofresh/action-changed-files@v3
+ id: metadata
+ with:
+ pattern: docker/(?P\w+)/.*
+ default-patterns: |
+ meta.yml
+ Dockerfile
+
+ update-docker:
+ needs: [generate-matrix]
+ strategy:
+ matrix: ${{ fromJson(needs.generate-matrix.outputs.matrix-metadata) }}
+ if: ${{ fromJson(needs.generate-matrix.outputs.matrix-metadata).include[0] }} # skip if the matrix is empty!
+ runs-on: ubuntu-latest
+ steps:
+ - uses: actions/checkout@v4
+ - uses: pietrobolcato/action-read-yaml@1.0.0
+ id: metadata
+ with:
+ config: ${{ github.workspace }}/docker/${{ matrix.image_dir }}/meta.yml
+ - name: Get date
+ id: date
+ run: |
+ echo "DATE=$(date +"%Y-%m-%d")" >> $GITHUB_OUTPUT
+ - name: Login to DockerHub
+ if: github.event_name != 'pull_request'
+ uses: docker/login-action@v2
+ with:
+ username: ${{ secrets.DOCKERHUB_USERNAME }}
+ password: ${{ secrets.DOCKERHUB_TOKEN }}
+ - name: Build and push
+ uses: docker/build-push-action@v4
+ # only try building & pushing the container if parsing the metadata worked
+ if: ${{ steps.metadata.outputs['container'] != '' }}
+ with:
+ context: docker/${{ matrix.image_dir }}
+ # only push container to docker hub if not triggered from a PR
+ push: ${{ github.event_name != 'pull_request' }}
+ tags: ${{ steps.metadata.outputs['container'] }}
+ build-args: |
+ BUILD_DATE=${{ steps.date.outputs.DATE }}
+ BUILD_TAG=${{ steps.metadata.outputs['version'] }}
+ REPONAME=${{ steps.metadata.outputs['image_name'] }}
diff --git a/CHANGELOG.md b/CHANGELOG.md
index 92263d0..d42f36c 100644
--- a/CHANGELOG.md
+++ b/CHANGELOG.md
@@ -1,7 +1,16 @@
## XAVIER development version
+- Fix minor bug in assess_significance.R script associated with rule freec_exome_somatic (#120, @samarth8392)
- Fix `cache` subcommand to correctly read the container images config file. (#124, @kelly-sovacool)
+## XAVIER 3.1.1
+
+- New contributing guide available on GitHub and the documentation website. (#114, @kelly-sovacool)
+- New `xavier debug` subcommand to determine the base directory for debugging purposes. (#114, @kelly-sovacool)
+- Upgraded `ccbr_wes_base` docker to v1.1.0 with updated GATK version to v4.6.0.0 (#116, @samarth8392)
+- Upgrade multiqc container to use v1.15. (#117, @kelly-sovacool)
+- Upgrade memory for rule "bwa_mem" to 100G (#118, @samarth8392)
+
## XAVIER 3.1.0
### new features
diff --git a/CITATION.cff b/CITATION.cff
index c8f815c..ecee53e 100644
--- a/CITATION.cff
+++ b/CITATION.cff
@@ -46,5 +46,5 @@ identifiers:
- description: Archived snapshots of all versions
type: doi
value: 10.5281/zenodo.12727315
-version: v3.1.0
-date-released: "2024-08-27"
+version: v3.1.1
+date-released: "2024-10-02"
diff --git a/VERSION b/VERSION
index 0f9d6b1..8d70082 100644
--- a/VERSION
+++ b/VERSION
@@ -1 +1 @@
-3.1.0-dev
+3.1.1-dev
diff --git a/config/cluster.biowulf.json b/config/cluster.biowulf.json
index a647265..7856824 100644
--- a/config/cluster.biowulf.json
+++ b/config/cluster.biowulf.json
@@ -120,7 +120,7 @@
},
"bwa_mem": {
"threads": "24",
- "mem": "64G"
+ "mem": "100G"
},
"picard_headers": {
"threads": "2",
diff --git a/config/cluster.frce.json b/config/cluster.frce.json
index 6c7773d..1fe95e1 100644
--- a/config/cluster.frce.json
+++ b/config/cluster.frce.json
@@ -119,7 +119,7 @@
},
"bwa_mem": {
"threads": "24",
- "mem": "64G"
+ "mem": "100G"
},
"picard_headers": {
"threads": "2",
diff --git a/config/containers/images.json b/config/containers/images.json
index 7c35a96..68f2ddd 100644
--- a/config/containers/images.json
+++ b/config/containers/images.json
@@ -1,6 +1,6 @@
{
"images": {
- "wes_base": "docker://nciccbr/ccbr_wes_base:v0.1.0",
+ "wes_base": "docker://nciccbr/ccbr_wes_base:1.1.0",
"vcf2maf": "docker://dnousome/ccbr_vcf2maf:v102.0.0",
"mutect": "docker://nciccbr/ccbr_mutect:v0.1.0",
"fastq_screen": "docker://nciccbr/ccbr_fastq_screen_0.13.0:v2.0",
@@ -8,7 +8,7 @@
"fastqvalidator": "docker://nciccbr/ccbr_fastqvalidator:v0.1.0",
"kraken": "docker://nciccbr/ccbr_kraken_v2.1.1:v0.0.1",
"miniconda": "docker://continuumio/miniconda3:4.9.2",
- "multiqc": "docker://nciccbr/ccbr_multiqc_1.9:v0.0.1",
+ "multiqc": "docker://nciccbr/ccbr_multiqc_1.15:v1",
"picard": "docker://nciccbr/ccbr_picard:v0.0.1",
"python": "docker://nciccbr/ccbr_python:v0.0.1",
"qualimap": "docker://nciccbr/ccbr_qualimap:v0.0.1"
diff --git a/docker/wes_base/Dockerfile b/docker/wes_base/Dockerfile
index 209eadd..dff757b 100644
--- a/docker/wes_base/Dockerfile
+++ b/docker/wes_base/Dockerfile
@@ -1,7 +1,14 @@
# Base image
-FROM ubuntu:20.04
+FROM nciccbr/ccbr_ubuntu_base_20.04:v6
-MAINTAINER
+ARG BUILD_DATE="000000"
+ENV BUILD_DATE=${BUILD_DATE}
+ARG BUILD_TAG="000000"
+ENV BUILD_TAG=${BUILD_TAG}
+ARG REPONAME="000000"
+ENV REPONAME=${REPONAME}
+
+LABEL maintainer
# Create Container filesystem specific
# working directory and opt directories
@@ -38,11 +45,12 @@ RUN apt-get update \
python3 \
python3-pip \
unzip \
- wget
+ wget \
+ less
# Make python3 the default interpreter
# and install Python Packages
-RUN ln -s /usr/bin/python3.8 /usr/bin/python
+#RUN ln -s /usr/bin/python3.8 /usr/bin/python
RUN pip3 install --upgrade pip \
&& pip3 install argparse \
&& pip3 install numpy \
@@ -89,7 +97,14 @@ RUN DEBIAN_FRONTEND=noninteractive apt-get install -y \
manpages-dev \
zlib1g \
zlib1g-dev \
- zlibc
+ zlibc \
+ openjdk-17-jdk \
+ openjdk-11-jdk
+
+# Install Java 17
+# RUN apt-get update && apt-get install -y openjdk-17-jdk
+# ENV JAVA_HOME=/usr/lib/jvm/java-17-openjdk-amd64
+# ENV PATH="$JAVA_HOME/bin:${PATH}"
# Common bioinformatics tools
# bwa/0.7.17 bowtie/1.2.3 bowtie2/2.3.5.1
@@ -115,13 +130,13 @@ RUN wget https://github.com/biod/sambamba/releases/download/v0.8.1/sambamba-0.8.
&& mv /opt2/sambamba-0.8.1-linux-amd64-static /opt2/sambamba \
&& chmod a+rx /opt2/sambamba
-# Install GATK4 (GATK/4.2.2.0)
-# Requires Java8 or 1.8
-RUN wget https://github.com/broadinstitute/gatk/releases/download/4.2.2.0/gatk-4.2.2.0.zip \
- && unzip /opt2/gatk-4.2.2.0.zip \
- && rm /opt2/gatk-4.2.2.0.zip \
- && /opt2/gatk-4.2.2.0/gatk --list
-ENV PATH="/opt2/gatk-4.2.2.0:$PATH"
+# Install GATK4 (GATK/4.6.0.0)
+# Requires Java17
+RUN wget https://github.com/broadinstitute/gatk/releases/download/4.6.0.0/gatk-4.6.0.0.zip \
+ && unzip /opt2/gatk-4.6.0.0.zip \
+ && rm /opt2/gatk-4.6.0.0.zip \
+ && /opt2/gatk-4.6.0.0/gatk --list
+ENV PATH="/opt2/gatk-4.6.0.0:$PATH"
# Install last release of GATK3 (GATK/3.8-1)
# Only being used for the CombineVariants
@@ -175,12 +190,12 @@ RUN pip3 install --upgrade pip \
RUN wget https://github.com/brentp/somalier/releases/download/v0.2.13/somalier \
&& chmod a+rx /opt2/somalier
-# Install VarScan/v2.4.4
+# Install VarScan/v2.4.6
# Works with java8
# and each wrapper script similar to HPC module
-RUN wget https://github.com/dkoboldt/varscan/raw/master/VarScan.v2.4.4.jar \
+RUN wget https://github.com/dkoboldt/varscan/raw/master/VarScan.v2.4.6.jar \
&& echo '#!/bin/bash' > /opt2/varscan \
- && echo 'java -jar /opt2/VarScan.v2.4.4.jar "$@"' >> /opt2/varscan \
+ && echo 'java8 -jar /opt2/VarScan.v2.4.6.jar "$@"' >> /opt2/varscan \
&& chmod a+rx /opt2/varscan
# Install snpEff/4.3t
@@ -195,7 +210,7 @@ ENV SNPEFF_JAR="/opt2/snpEff/snpEff.jar"
# Install samblaster/0.1.26
# Requires g++ and make (already satisfied)
-RUN git clone git://github.com/GregoryFaust/samblaster.git \
+RUN git clone https://github.com/GregoryFaust/samblaster.git \
&& cd /opt2/samblaster \
&& make \
&& chmod a+rX /opt2/samblaster/samblaster
@@ -234,7 +249,7 @@ ENV PATH="/opt2/VarDict-1.8.2/bin:$PATH"
# and set java8 as default with links
# to alternative versions
ADD Dockerfile /opt2/base_gatk4_wes.dockerfile
-COPY argparse.bash /opt2
+COPY /argparse.bash /opt2
# Trimmomatic requires java11
ENV TRIMMOMATIC_JAR="/usr/share/java/trimmomatic-0.39.jar"
RUN echo '#!/bin/bash' > /opt2/trimmomatic \
@@ -242,9 +257,10 @@ RUN echo '#!/bin/bash' > /opt2/trimmomatic \
&& chmod +x /opt2/trimmomatic
RUN chmod -R a+rX /opt2 \
&& chmod a+x /opt2/argparse.bash \
+ && ln -s /usr/lib/jvm/java-17-openjdk-amd64/bin/java /usr/bin/java17 \
&& ln -s /usr/lib/jvm/java-11-openjdk-amd64/bin/java /usr/bin/java11 \
&& ln -s /usr/lib/jvm/java-8-openjdk-amd64/bin/java /usr/bin/java8 \
- && ln -fs /usr/lib/jvm/java-8-openjdk-amd64/bin/java /usr/bin/java
+ && ln -fs /usr/lib/jvm/java-17-openjdk-amd64/bin/java /usr/bin/java
ENV PATH="/opt2:$PATH"
ENV TRIMMOJAR="/usr/share/java/trimmomatic-0.39.jar"
@@ -260,3 +276,6 @@ RUN DEBIAN_FRONTEND=noninteractive apt-get install -y \
pandoc \
&& apt-get clean && apt-get purge \
&& rm -rf /var/lib/apt/lists/* /tmp/* /var/tmp/*
+
+COPY Dockerfile /opt2/Dockerfile_${REPONAME}.${BUILD_TAG}
+RUN chmod a+r /opt2/Dockerfile_${REPONAME}.${BUILD_TAG}
\ No newline at end of file
diff --git a/docker/wes_base/meta.yml b/docker/wes_base/meta.yml
new file mode 100644
index 0000000..5a252c9
--- /dev/null
+++ b/docker/wes_base/meta.yml
@@ -0,0 +1,4 @@
+dockerhub_namespace: nciccbr
+image_name: ccbr_wes_base
+version: 1.1.0
+container: "$(dockerhub_namespace)/$(image_name):$(version)"
diff --git a/docs/contributing.md b/docs/contributing.md
new file mode 100644
index 0000000..773a7e5
--- /dev/null
+++ b/docs/contributing.md
@@ -0,0 +1,3 @@
+--8<-- ".github/CONTRIBUTING.md"
+
+
diff --git a/mkdocs.yml b/mkdocs.yml
index 130fb76..b61d5a6 100644
--- a/mkdocs.yml
+++ b/mkdocs.yml
@@ -113,4 +113,5 @@ nav:
- Output Files: pipeline-details/output.md
- FAQ:
- General Questions: faq/questions.md
+ - How to contribute: contributing.md
- License: license.md
diff --git a/pyproject.toml b/pyproject.toml
index 3e53d62..0f12601 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -42,7 +42,6 @@ dependencies = [
"Click >= 8.1.3",
"PySimpleGui < 5",
"snakemake >= 7.32, < 8",
- "snaketool-utils >= 0.0.5",
]
[project.optional-dependencies]
diff --git a/src/xavier/__main__.py b/src/xavier/__main__.py
index 621fcfe..fdf1761 100755
--- a/src/xavier/__main__.py
+++ b/src/xavier/__main__.py
@@ -658,8 +658,15 @@ def parsed_arguments():
help="Only display what remote resources would be pulled.",
)
+ subparser_debug = subparsers.add_parser(
+ "debug",
+ help="Debug the pipeline base directory.",
+ usage=argparse.SUPPRESS,
+ )
+
# Define handlers for each sub-parser
subparser_run.set_defaults(func=run)
+ subparser_debug.set_defaults(func=debug)
subparser_unlock.set_defaults(func=unlock)
subparser_cache.set_defaults(func=cache)
subparser_gui.set_defaults(func=launch_gui)
@@ -669,6 +676,11 @@ def parsed_arguments():
return args
+def debug(args):
+ print("BASE:", xavier_base())
+ print(get_version(debug=True))
+
+
def main():
# show helpful error message when no arguments given
if len(sys.argv) == 1:
diff --git a/src/xavier/util.py b/src/xavier/util.py
index 4f04bfa..d62a338 100644
--- a/src/xavier/util.py
+++ b/src/xavier/util.py
@@ -22,10 +22,13 @@ def xavier_base(*paths):
return str(basedir.joinpath(*paths))
-def get_version():
+def get_version(debug=False):
"""Get the current version
@return version
"""
- with open(xavier_base("VERSION"), "r") as vfile:
+ version_file = xavier_base("VERSION")
+ if debug:
+ print("VERSION FILE:", version_file)
+ with open(version_file, "r") as vfile:
version = f"v{vfile.read().strip()}"
return version
diff --git a/tests/test_cli.py b/tests/test_cli.py
index 2196e3f..99abb44 100644
--- a/tests/test_cli.py
+++ b/tests/test_cli.py
@@ -33,6 +33,11 @@ def test_help():
assert "XAVIER" in shell_run("./bin/xavier --help")
+def test_debug():
+ out = shell_run("./bin/xavier debug")
+ assert all(["xavier (v" in out, "BASE: /" in out, "VERSION FILE: /" in out])
+
+
def test_dryrun_targets():
if get_hpcname() == "biowulf":
output_human, config_human = run_in_temp(f"{xavier_run} --genome hg38")
diff --git a/workflow/rules/qc.smk b/workflow/rules/qc.smk
index b5f1eaa..1e45b6c 100644
--- a/workflow/rules/qc.smk
+++ b/workflow/rules/qc.smk
@@ -390,7 +390,7 @@ rule snpeff:
envmodules: config['tools']['snpEff']['modname']
container: config['images']['wes_base']
shell: """
- java -Xmx12g -jar $SNPEFF_JAR \\
+ java8 -Xmx12g -jar $SNPEFF_JAR \\
-v -canon -c {params.config} \\
-csvstats {output.csv} \\
-stats {output.html} \\
diff --git a/workflow/rules/somatic_snps.common.smk b/workflow/rules/somatic_snps.common.smk
index 27e6989..121b2c7 100644
--- a/workflow/rules/somatic_snps.common.smk
+++ b/workflow/rules/somatic_snps.common.smk
@@ -168,7 +168,7 @@ rule somatic_merge_callers:
mkdir -p "$(dirname {output.mergedvcf})"
fi
- java -Xmx60g -Djava.io.tmpdir=${{tmp}} -jar $GATK_JAR -T CombineVariants \\
+ java8 -Xmx60g -Djava.io.tmpdir=${{tmp}} -jar $GATK_JAR -T CombineVariants \\
-R {params.genome} \\
-nt {threads} \\
--filteredrecordsmergetype KEEP_IF_ANY_UNFILTERED \\
diff --git a/workflow/rules/somatic_snps.paired.smk b/workflow/rules/somatic_snps.paired.smk
index 1f5427b..42e902e 100644
--- a/workflow/rules/somatic_snps.paired.smk
+++ b/workflow/rules/somatic_snps.paired.smk
@@ -147,7 +147,7 @@ rule strelka:
cd "$myoutdir"
./runWorkflow.py -m local -j {threads}
- java -Xmx12g -Djava.io.tmpdir=${{tmp}} -XX:ParallelGCThreads={threads} \\
+ java8 -Xmx12g -Djava.io.tmpdir=${{tmp}} -XX:ParallelGCThreads={threads} \\
-jar $GATK_JAR -T CombineVariants \\
-R {params.genome} \\
--variant results/variants/somatic.snvs.vcf.gz \\
@@ -439,7 +439,7 @@ rule varscan_paired:
awk '{{gsub(/\y[W|K|Y|R|S|M|B|D|H|V]\y/,"N",$4); OFS = "\t"; print}}' {output.vcf}.indel \\
| sed '/^$/d' > {output.vcf}.indel_temp
- java -Xmx12g -Djava.io.tmpdir=${{tmp}} -XX:ParallelGCThreads={threads} \\
+ java8 -Xmx12g -Djava.io.tmpdir=${{tmp}} -XX:ParallelGCThreads={threads} \\
-jar $GATK_JAR -T CombineVariants \\
-R {params.genome} \\
--variant {output.vcf}.snp_temp \\
diff --git a/workflow/scripts/assess_significance.R b/workflow/scripts/assess_significance.R
index e32f7f7..3e64bc4 100644
--- a/workflow/scripts/assess_significance.R
+++ b/workflow/scripts/assess_significance.R
@@ -48,7 +48,7 @@ for (i in c(1:length(cnvs[, 1]))) {
}
KS <- function(values, normals) {
resultks <- try(ks.test(values, score(normals)), silent = TRUE)
- if (class(resultks) == "try-error") {
+ if (paste(class(resultks), collapse="_") == "try-error") {
return(list("statistic" = NA, "p.value" = NA, "alternative" = NA, "method" = NA, "data.name" = NA))
} else {
resultks