-
Notifications
You must be signed in to change notification settings - Fork 978
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Faster uniqueN(v) for logical v #2648
Conversation
Codecov Report
@@ Coverage Diff @@
## master #2648 +/- ##
==========================================
+ Coverage 93.17% 93.32% +0.15%
==========================================
Files 61 61
Lines 12169 12427 +258
==========================================
+ Hits 11338 11598 +260
+ Misses 831 829 -2
Continue to review full report at Codecov.
|
|
this is nice. I feel like a more custom-tailored version could do even Q about your examples: looks like The worst case for what I have in mind would be something like This is hard for |
|
Thanks, Michael. Yes, you're right that it would be better to avoid 2 or 3 passes over the vector. I don't have the skill to write a C function that could outperform base R's. Maybe I should learn how to, but I think it's best not to practise on you guys! That being said, if the unique values aren't too clustered as they are in the worst case, Re: worst case, |
|
yes indeed. and i think PR should be approved. just making note that further improvement can happen "down the line" |
|
Great idea! I couldn't resist doing it in C. Single pass and returns early. Its logic can probably be simplified, needs more eyes to check please. Maybe this can be used as a template example of how to add a fairly simple C function to the package. |
src/uniqlist.c
Outdated
| while (i<n && INTEGER(x)[i]==first) i++; | ||
| if (i==n) | ||
| return( ScalarInteger( first==NA_INTEGER && narm ? 0 : 1) ); // all one value | ||
| Rboolean second = INTEGER(x)[i]; |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
@mattdowle if na.rm, we can short-circuit here if both first and second are not NA_INTEGER
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
it already does that, iiuc, on line 253 with the comment // TRUE and FALSE found before any NA, but na.rm=TRUE so we're done
|
@mattdowle indeed, the code is about as straightforward as it comes from C. Good to see an isolated example like this, makes it clearer all the necessary steps to not just write the code but register it for compilation & use in R. |
|
Strange. I'm getting slower speeds for |
|
This is what I just got on the first attempt: very small timings and the first one is 14ms vs the subsequent at 4ms. Could something like that have happened (one relative outlier)? What IIUC, |
…just simpler code.
…hird started. No speed difference, just tidying.
|
Can't think of any other refinements. Will wait for @HughParsonage to confirm the 7% figure before merging. |
|
I'm using It might be something to do with
and yes |
|
Interesting! I tried to reproduce but I see a much smaller difference. When you test the dev version of data.table are you compiling with -O3? [ All just for fun since difference is so small. ] So I see a 0.5ms difference (<1%) here, where you're seeing 3ms - 14ms (4% - 12%). |
|
Ah, no |
|
Worth trying -O3 then for fun. It could be branch prediction, too. Looking at base R, it uses a for loop and an That's giving the compiler/cpu pipeline a better chance to run ahead. We don't care where it finds the NA_LOGICAL just whether it does or not. So that might be helping it in base R. |
|
Hmm, very noob problem, but setting the optimization flag to
(Don't worry if this will take too long.) |
|
NP. Try passing |
|
Ah so close |
|
Try: haskell/unix#49 |
|
Oh were it that simple on Windows... I tried the binary version via |
|
Ok, please give that a go. Installing the AppVeyor binary is a good call. Installing the package (rather than Also make sure that nothing else is running on your machine. Chrome is especially heavy. On Windows I don't recall if system.time() distinguishes user + system != elapsed. And I don't know if microbenchmark uses use+system as it should, or elapsed. Elapsed (wall clock time) is affected by other processes on the machine. Especially at this tiny scale! So I close Chrome, Slack, Pandora etc. Or run on a server. The high |
|
Nope: |
|
Can you try : |
> microbenchmark(uniqueN(rand_2), unique_N(rand_2), control=list(order="block"))
Unit: milliseconds
expr min lq mean median uq max neval cld
uniqueN(rand_2) 81.38844 81.6229 83.15188 81.80060 82.10328 154.4352 100 b
unique_N(rand_2) 76.48197 76.6827 77.83888 76.81929 77.18733 102.8325 100 a |
|
How odd. I reproduced on Windows too. > microbenchmark(uniqueN(rand_2), unique_N(rand_2), control=list(order="block"))
Unit: milliseconds
expr min lq mean median uq max neval
uniqueN(rand_2) 51.16815 51.94226 53.48157 52.37371 53.14368 87.33350 100
unique_N(rand_2) 49.37339 49.95491 50.82088 50.18038 50.52716 82.32456 100 |
|
The AppVeyor logs show it is using -O2. I tried with -O2 locally but I see each as the same timing, unlike on Windows. I'm at a loss then. |
|
My instinct is to go with base R only to keep the compiling to a minimum. Should keep the test suite of course. But the bulk of the gain from 1.10.4-3 has been achieved either way. happy to improve my other PR :-) btw |
|
That last commit resulted in a smaller gap: |
|
That seems better on my Windows VM : > microbenchmark(uniqueN(rand_2), unique_N(rand_2), control=list(order="block"))
Unit: milliseconds
expr min lq mean median uq max neval
uniqueN(rand_2) 48.78577 49.35803 50.02199 49.76901 50.29283 60.40149 100
unique_N(rand_2) 48.68613 49.42341 50.61808 49.82927 50.42004 58.82965 100 |
|
Ok great. That makes sense then. int (32bit) type on a 64bit cpu has weird effects. It was the last thing that was different to base R. Interesting it helps a bit more on Windows. Good to discover. With 5e8 on Windows VM : > N = 5e8
> rand_2 <- sample(c(TRUE, FALSE), size = N, replace = TRUE)
> microbenchmark(uniqueN(rand_2), unique_N(rand_2), control=list(order="block"))
Unit: milliseconds
expr min lq mean median uq max neval
uniqueN(rand_2) 241.2369 247.9918 253.8572 249.4949 253.8268 292.5559 100
unique_N(rand_2) 241.3590 248.2655 254.1266 249.9260 254.8916 287.5942 100 |
|
What's the comparison for all of the tests, now? If the |
|
Where does that leave us across all tests? |
|
(And probably faster for the |
|
LGTM |
|
This case shows the double pass I had in mind. (Which I now realize is covered by the chart!) > N = 1e8
> end = c(rep(TRUE,N),FALSE,NA)
> uniqueN(end)
[1] 3
> unique_N(end)
[1] 3
> microbenchmark(uniqueN(end), unique_N(end), control=list(order="block"))
Unit: milliseconds
expr min lq mean median uq max neval
uniqueN(end) 49.81174 50.12209 51.06473 50.33556 50.69376 87.14682 100
unique_N(end) 99.37851 100.19555 101.59117 100.76662 101.57558 120.54343 100
> end = c(rep(NA,N),TRUE,FALSE)
> uniqueN(end)
[1] 3
> unique_N(end)
[1] 3
> microbenchmark(uniqueN(end), unique_N(end), control=list(order="block"))
Unit: milliseconds
expr min lq mean median uq max neval
uniqueN(end) 48.80704 49.28497 49.81357 49.62486 50.15754 56.29367 100
unique_N(end) 109.08013 110.32095 112.15381 110.96922 111.82071 131.69756 100 |
|
I did start to regret biting on this one. But the |
|
But what I did here in C is negligible compared to Hugh's original which for vast majority of real-world cases is an infinite improvement and unbeatable. I've put a simplified version of this into NEWS to convey how big an improvement it is. [ A few other news items could do with similar timings adding just to convey the order of the improvement, very briefly. ] |
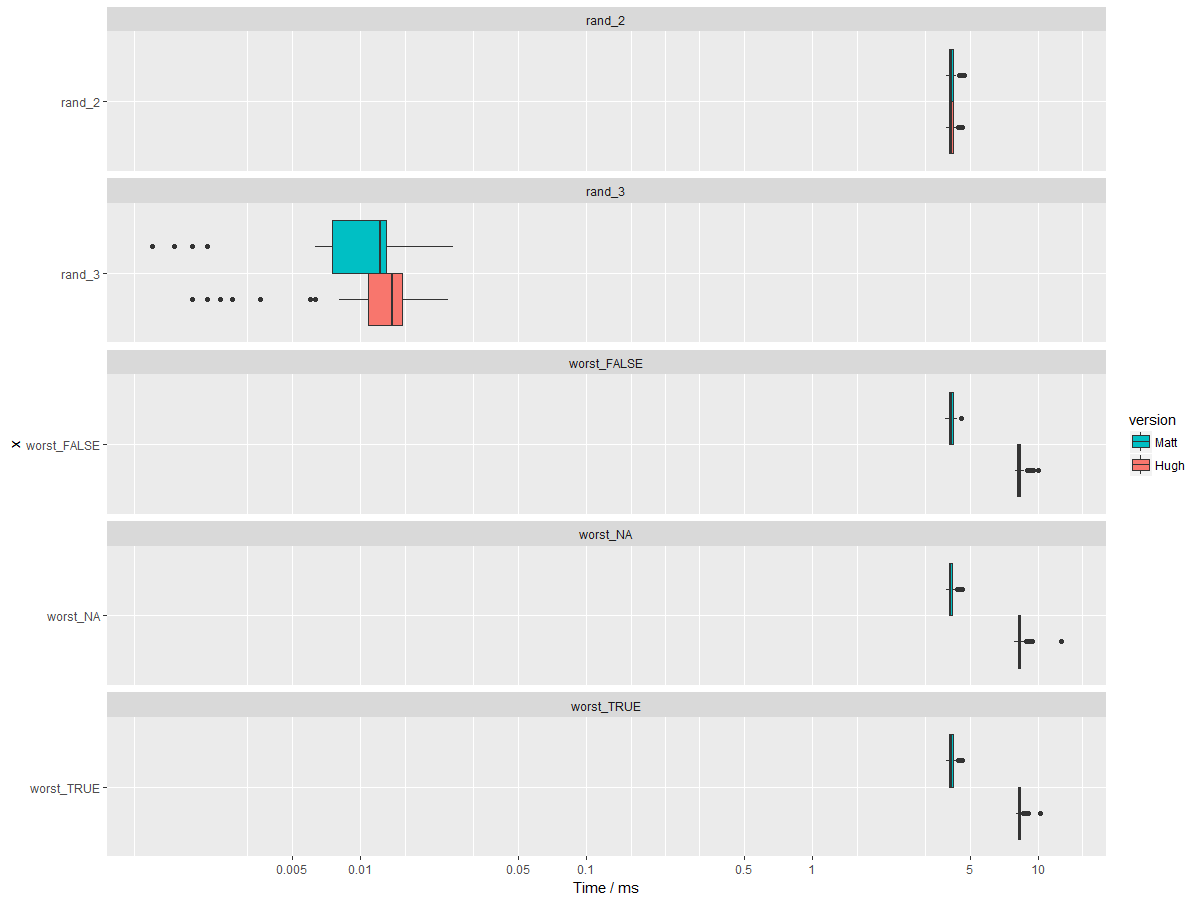
For
uniqueN(v)whenvis a logical, atomic vector, we can utilizeanyNA,anyandallfor a faster answer, especially whenanyNA(v). On a 30Mdata.tablewith 50 logical columns,vapply(DT, uniqueN, logical(1L))experienced a 20 s speedup (from 50s to 30s).The only downside is that
is.logicalhas to be run, though that is a trivial cost (could not be measured bymicrobenchmark).Additional benchmarks (I've entered what I consider to be worst-case scenarios for the new version, though others may be able to find worse instances).