BD2KPubMed is a tool to identify trends in the literature, quantifying the impact of particular proteins on current and future research.
BD2KPubMed is a software tool that employs publicly available data to analyze scientific trends and research directions in a particular field of interest. The software collects metadata associated with individual articles from a custom query of the NCBI PubMed database, then ranks proteins by their occurrences in the referenced articles. The data are then summarized for visualization and trend analysis and quantify the impact of a particular protein to a research topic. Potential applications include gathering information to prioritize in-depth gene annotation and mass spectrometry assay development.
- Download the latest version of the gene2pubmed file from the NCBI Gene database using the following link: ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/gene2pubmed.gz
- Use an file archiving tool such as 7zip to extract the gene2pubmed.gz file. Put the extracted contents of this file in a folder.
-
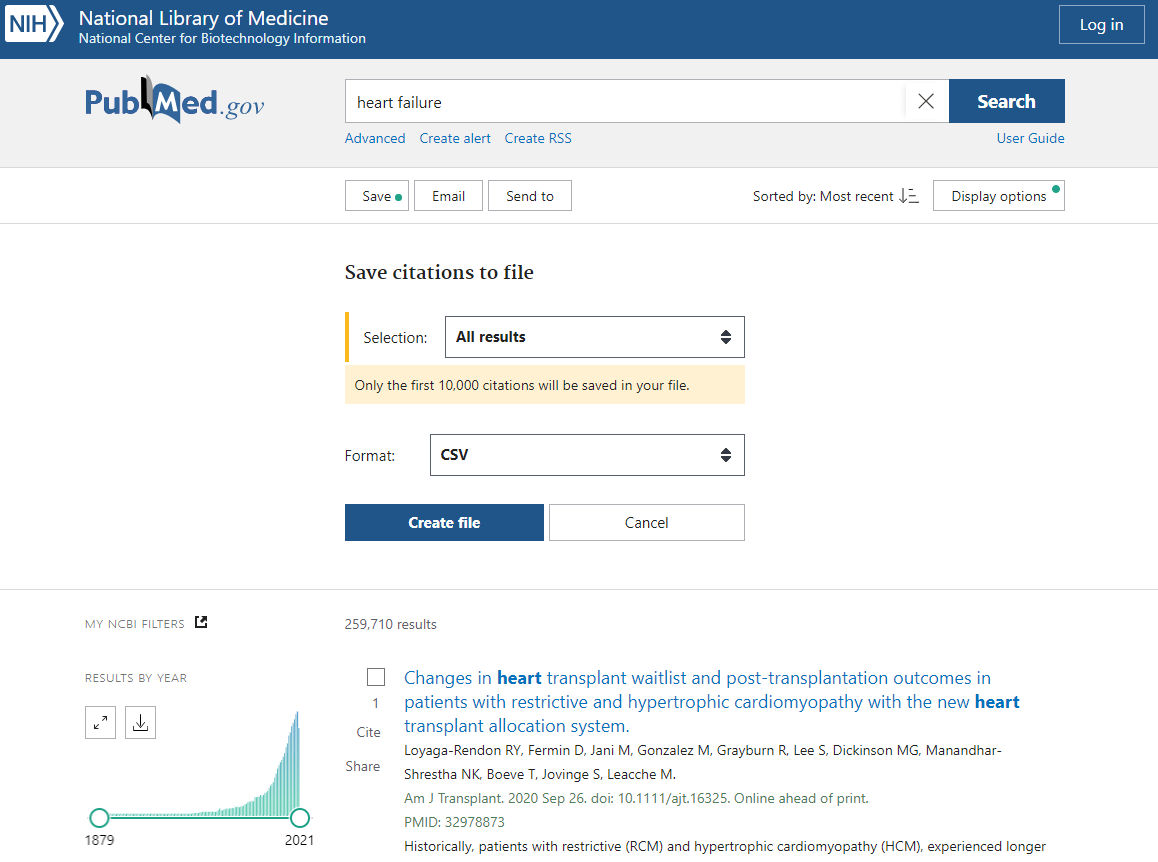
Make a PubMed search in the topic you are interested in. Above we give an example for the query “heart failure”.
- Click the “Save” button
- Choose your selection. Note that "all results" has a maximum of 10,000 citations. For more, download citations through the Entrez PubMed API.
- Choose the format “CSV” format
- Click “Create File”. Download this file into the same folder that contains the extracted gene2pubmed file.
-
Run the BD2KPubMed application by executing the BD2KPubMed.jar file in the dist folder. If you get an error, it is likely that you do not have the Java Runtime Environment installed on your system. You can install it on https://java.com/en/download/.
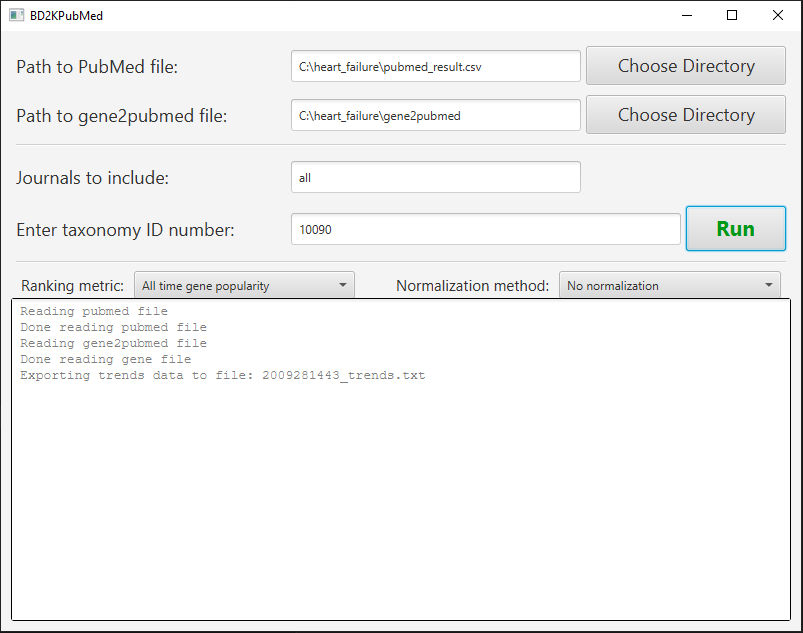
- Fill out the parameters for the analysis.
- Click the first Choose Directory button and select the .csv file downloaded from PubMed for the first field.
- Click the second Choose Directory button and select the extracted gene2pubmed file downloaded from the NCBI Gene database.
- Enter the journals you wish to include in your search, separated by a comma. You may type “all” or just leave this field blank to search all available journals.
- Type the taxonomy ID number of the organism you are interested in (e.g. 9606 for Human and 10090 for Mouse.)
- Choose your ranking metric.
- Select “All time gene popularity” to rank the genes by the total number of articles studying them.
- Select “Last 5 years gene popularity” to rank the genes by the total number of articles published in the last 5 years studying them.
- Select “Last 10 years gene popularity” to rank the genes by the total number of articles published in the last 10 years studying them.
- Choose your normalization method.
- Select “No normalization” for no normalization.
- Select “By gene reference” to normalize the gene popularities by the total number of gene citations for your topic that year. For example, if a specific gene is mentioned in 12 of the 1000 publications published in year 2011 and related to your PubMed search query, the output will be .0012 for this gene at the year 2011. (Please note that this kind of normalization tends to produce very high percentages for earlier years (e.g. 1970s), when the total gene citation output is very low, and the gene in question could be one of the few genes studied in that field that year that yielded to a publication. For highest quality results, avoid the use of the “All time gene popularity” option while using by gene reference normalization.)
- Click Run. The tool will provide up to 3 different outputs in the same location as the application:
- The popularity heat map will show the most popular genes in darker shades. The vertical axis represents the individual genes while the horizontal axis denotes the year.
- The trends text file in the same folder as the jar file. This contains the heatmap data in textual format.
- The gene_frequencies text file in the same folder as the jar file. This file contains the sum of number of publications mentioning the particular gene, sorted by metric.
For questions, concerns, and technical support, please open an issue on this GitHub repository.