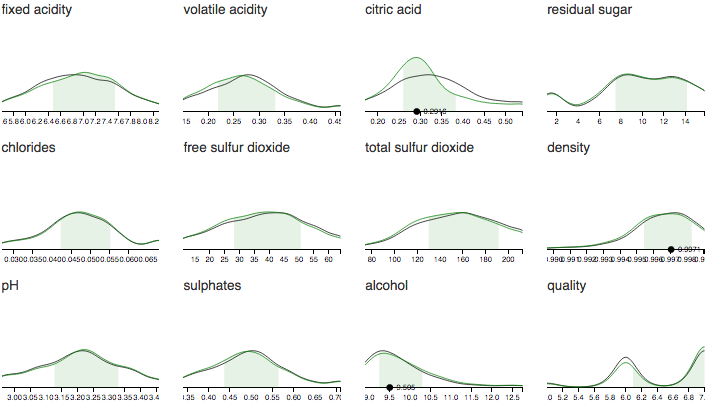
An experimental model for robust dimensionality reduction of arbitrary data sets. It lets you explore the marginal distributions of all parameters with any parameter(s) fixed. It is also extremely fast at computing these probability distributions. It assumes nothing about the distribution of the features, which can have any units and have any scale.
- Predicting missing values
- Visualizing probability distributions
- Finding outliers
- Modelling uncertainties
Here is a demo trained on a bunch of different data sets. The demo is written in JS and uses pre-trained models.
Tools like Crossfilter are great at visualizing datasets and how features are correlated. Crossfilter renders real data points based on a number of feature selections. However as the number of features increase, it gets harder to find data points that fulfill all criteria. This is the curse of dimensionality problem which often makes analysis of high-dimensional data hard.
This package has a different approach. It computes a statistical model of the underlying data. The downside is that you can no longer explore real data points. The upside is you can explore conditional dependencies and make predictions about data you have not observed.
It builds an autoencoder that learns to reconstruct missing data.
To be able to work with any distributions, it reduces all inputs to a series of binary values. Every feature is encoded as a binary feature vector by constructing splits from the empirical distribution of the training data. The predicted probabilities for each split then gives the CDF directly.
The autoencoder has a series of hidden bottleneck layers (typically 2-5 layers with 20-100 units). One way to think of it is that the autoencoder finds a low-dimensional manifold in the high-dimensional space. This manifold can be highly nonlinear due to the nonlinearities in the autoencoder. The autoencoder then essentially learns a projection from the high dimensional space onto the manifold and another projection back to the original space. The dimensionality reduction is effectively a way of getting around the curse of dimensionality.
The autoencoder is trained using Theano in Python. You can run it on a GPU although the speed improvements are not drastic because of some bottlenecks.
- Training the autoencoder is very slow. It can take a few hours for small data sets (with 10-100 features).
- The training time grows with the number of features.
- The training time should stay relatively constant with the size of the training data since it is using stochastic gradient descent.
- It only handles numerical attributes, although categorical should be pretty easy to add.
- The model probably overfits quite a bit and should do proper cross-validation to measure this.
- Similarly there is a bunch of hyperparameters that have to be tuned to your data set.
Released under the Apache 2.0 license.