pymia is an open-source Python (py) package for deep learning-based medical image analysis (mia). The package addresses two main parts of deep learning pipelines: data handling and evaluation. The package itself is independent of the deep learning framework used but can easily be integrated into TensorFlow and PyTorch pipelines. Therefore, pymia is highly flexible, allows for fast prototyping, and reduces the burden of implementing data handling and evaluation.
The main features of pymia are data handling (pymia.data package) and evaluation (pymia.evaluation package).
The data package is used to extract data (images, labels, demography, etc.) from a dataset in the desired format (2-D, 3-D; full- or patch-wise) for feeding to a neural network.
The output of the neural network is then assembled back to the original format before extraction, if necessary.
The evaluation package provides both evaluation routines as well as metrics to assess predictions against references.
Evaluation can be used both for stand-alone result calculation and reporting, and for monitoring of the training progress.
Further, pymia provides some basic image filtering and manipulation functionality (pymia.filtering package).
We recommend following our examples.
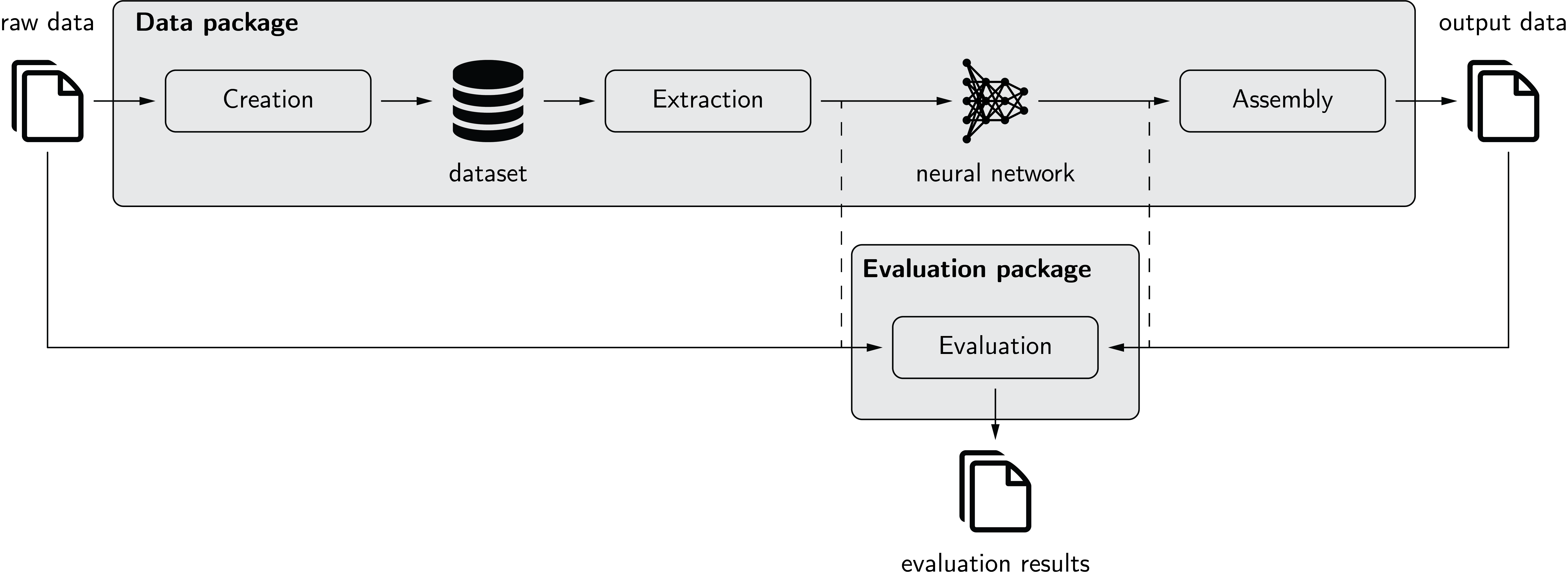
The following figure depicts pymia in the deep learning environment. The data package allows to create a dataset from raw data. Extraction of the data from this dataset is possible in nearly every desired format (2-D, 3-D; full- or patch-wise) for feeding to a neural network. The prediction of the neural network can, if necessary, be assembled back to the format before extraction. The evaluation package allows to evaluate predictions against references using a vast amount of metrics. It can be used stand-alone (solid) or for performance monitoring during training (dashed).
If you are new to pymia, here are a few guides to get you up to speed right away:
- Installation for installation instructions - or simply run
pip install pymia - Examples give you an overview of pymia's intended use. Jupyter notebooks and Python scripts are available in the directory ./examples.
- Do you want to contribute?
- Change history
- Acknowledgments
If you use pymia for your research, please acknowledge it accordingly by citing our paper:
BibTeX entry:
@article{Jungo2021a,
author = {Jungo, Alain and Scheidegger, Olivier and Reyes, Mauricio and Balsiger, Fabian},
doi = {10.1016/j.cmpb.2020.105796},
issn = {01692607},
journal = {Computer Methods and Programs in Biomedicine},
pages = {105796},
title = {{pymia: A Python package for data handling and evaluation in deep learning-based medical image analysis}},
volume = {198},
year = {2021},
}