Releases: rapidsai/cucim
v22.02.00
Version 22.02.00 (Feb 02, 2022)
This version would be available through both Conda (https://anaconda.org/rapidsai/cucim) and PyPI package (https://pypi.org/project/cucim/22.02.00/).
🚨 Breaking Changes
🐛 Bug Fixes
- Fix a bug in v21.12.01 (#191) @gigony
- Fix GPU memory leak when using nvJPEG API (when
device='cuda'parameter is used inread_regionmethod).
- Fix GPU memory leak when using nvJPEG API (when
- Fix segfault for preferred_memory_capacity in Python 3.9+ (#214) @gigony
📖 Documentation
🚀 New Features
- Update cucim.skimage API to match scikit-image 0.19 (#190) @glee77
- Support multi-threads and batch, and support nvJPEG for JPEG-compressed images (#191) @gigony
- Allow CuPy 10 (#195) @jakikham
1. Update cucim.skimage API to match scikit-image 0.19 (🚨 Breaking Changes)
channel_axis support
scikit-image 0.19 adds a channel_axis argument that should now be used instead of the multichannel boolean.
In scikit-image 1.0, the multichannel argument will likely be removed so we start supporting channel_axis in cuCIM.
This pulls changes from many scikit-image 0.19.0 PRs related to deprecating multichannel in favor of channel_axis. A few other minor PRs related to deprecations and updates to color.label2rgb are incorporated here as well.
The changes are mostly non-breaking, although a couple of deprecated functions have been removed (rgb2grey, grey2rgb) and a change in the default value of label2rgb's bg_label argument. The deprecated alpha argument was removed from gray2rgb.
Implements:
- Add saturation parameter to color.label2rgb #5156
- Decorators for helping with the multichannel->channel_axis transition #5228
- multichannel to channel_axis (1 of 6): features and draw #5284
- multichannel to channel_axis (2 of 6): transform functions #5285
- multichannel to channel_axis (3 of 6): filters #5286
- multichannel to channel_axis (4 of 6): metrics and measure #5287
- multichannel to channel_axis (5 of 6): restoration #5288
- multichannel to channel_axis (6 of 6): segmentation #5289
- channel_as_last_axis decorator fix #5348
- fix wrong error for metric.structural_similarity when image is too small #5395
- Add a channel_axis argument to functions in the skimage.color module #5462
- Remove deprecated functions and arguments for the 0.19 release #5463
- Support nD images and labels in label2rgb #5550
- remove need for channel_as_last_axis decorator in skimage.filters #5584
- Preserve backwards compatibility for
channel_axisparameter in transform functions #6095
Update float32 dtype support to match scikit-image 0.19 behavior
Makes float32 and float16 handling consistent with scikit-image 0.19. (must functions support float32, float16 gets promoted to float32)
Deprecate APIs
Introduces new deprecations as in scikit-image 0.19.
Specifically:
selem->footprintgrey->grayiterations->num_itermax_iter->max_num_itermin_iter->min_num_iter
2. Supporting Multithreading and Batch Processing
cuCIM now supports loading the entire image with multi-threads. It also supports batch loading of images.
If device parameter of read_region() method is "cuda", it loads a relevant portion of the image file (compressed tile data) into GPU memory using cuFile(GDS, GPUDirect Storage), then decompress those data using nvJPEG's Batched Image Decoding API.
Current implementations are not efficient and performance is poor compared to CPU implementations. However, we plan to improve it over the next versions.
Example API Usages
The following parameters would be added in the read_region method:
num_workers: number of workers(threads) to use for loading the image. (default:1)batch_size: number of images to load at once. (default:1)drop_last: whether to drop the last batch if the batch size is not divisible by the number of images. (default:False)preferch_factor: number of samples loaded in advance by each worker. (default:2)shuffle: whether to shuffle the input locations (default:False)seed: seed value for random value generation (default: 0)
Loading entire image by using multithreads
from cucim import CuImage
img = CuImage("input.tif")
region = img.read_region(level=1, num_workers=8) # read whole image at level 1 using 8 workersLoading batched image using multithreads
You can feed locations of the region through the list/tuple of locations or through the NumPy array of locations.
(e.g., ((<x for loc 1>, <y for loc 1>), (<x for loc 2>, <y for loc 2>)])).
Each element in the location should be int type (int64) and the dimension of the location should be
equal to the dimension of the size.
You can feed any iterator of locations (dimensions of the input don't matter, flattening the item in the iterator once if the item is also an iterator).
For example, you can feed the following iterator:
[0, 0, 100, 0]or(0, 0, 100, 0)would be interpreted as a list of(0, 0)and(100, 0).((sx, sy) for sy in range(0, height, patch_size) for sx in range(0, width, patch_size))would iterate over the locations of the patches.[(0, 100), (0, 200)]would be interpreted as a list of(0, 0)and(100, 0).- Numpy array such as
np.array(((0, 100), (0, 200)))ornp.array((0, 100, 0, 200))would be also available and using Numpy array object would be faster than using python list/tuple.
import numpy as np
from cucim import CuImage
cache = CuImage.cache("per_process", memory_capacity=1024)
img = CuImage("image.tif")
locations = [[0, 0], [100, 0], [200, 0], [300, 0],
[0, 200], [100, 200], [200, 200], [300, 200]]
# locations = np.array(locations)
region = img.read_region(locations, (224, 224), batch_size=4, num_workers=8)
for batch in region:
img = np.asarray(batch)
print(img.shape)
for item in img:
print(item.shape)
# (4, 224, 224, 3)
# (224, 224, 3)
# (224, 224, 3)
# (224, 224, 3)
# (224, 224, 3)
# (4, 224, 224, 3)
# (224, 224, 3)
# (224, 224, 3)
# (224, 224, 3)
# (224, 224, 3)Loading image using nvJPEG and cuFile (GDS, GPUDirect Storage)
If cuda argument is specified in device parameter of read_region() method, it uses nvJPEG with GPUDirect Storage to load images.
Use CuPy instead of Numpy, and Image Cache (CuImage.cache) wouldn't be used in the case.
import cupy as cp
from cucim import CuImage
img = CuImage("image.tif")
locations = [[0, 0], [100, 0], [200, 0], [300, 0],
[0, 200], [100, 200], [200, 200], [300, 200]]
# locations = np.array(locations)
region = img.read_region(locations, (224, 224), batch_size=4, device="cuda")
for batch in region:
img = cp.asarray(batch)
print(img.shape)
for item in img:
print(item.shape)
# (4, 224, 224, 3)
# (224, 224, 3)
# (224, 224, 3)
# (224, 224, 3)
# (224, 224, 3)
# (4, 224, 224, 3)
# (224, 224, 3)
# (224, 224, 3)
# (224, 224, 3)
# (224, 224, 3)Experimental Results
We have compared performance against Tifffile for loading the entire image.
System Information
- OS: Ubuntu 18.04
- CPU: Intel(R) Core(TM) i7-7800X CPU @ 3.50GHz, 12 processors.
- Memory: 64GB (G-Skill DDR4 2133 16GB X 4)
- Storage
- SATA SSD: Samsung SSD 850 EVO 1TB
Experiment Setup
Benchmarked loading several images with Tifffile.
- Use read_region() APIs to read the entire image (.svs/.tiff) at the largest resolution level.
- Performed on the following images that use a different compression method
- JPEG2000 YCbCr: TUPAC-TR-467.svs, 55MB, 19920x26420, tile size 240x240
- JPEG: image.tif (256x256 multi-resolution/tiled TIF conversion of TUPAC-TR-467.svs), 238MB, 19920x26420, tile size 256x256
- JPEG2000 RGB: [CMU-1-JP2K-33005.svs](https://opens...
- Performed on the following images that use a different compression method
v21.12.00
Version 21.12.00 (Dec 08, 2021)
This version would be available through both Conda (https://anaconda.org/rapidsai/cucim) and PyPI package (https://pypi.org/project/cucim/21.12.0/).
🚀 New Features
- Support Aperio SVS with CPU LZW and jpeg2k decoder (#141) @gigony
- Add NVTX support for performance analysis (#144) @gigony
- Normalize operation (#150) @shekhardw
1. Support Aperio SVS (.svs)
cuCIM now supports Aperio SVS format with help of OpenJpeg for decoding jpeg2k-compressed data.
Please check this notebook to see how to use the feature.
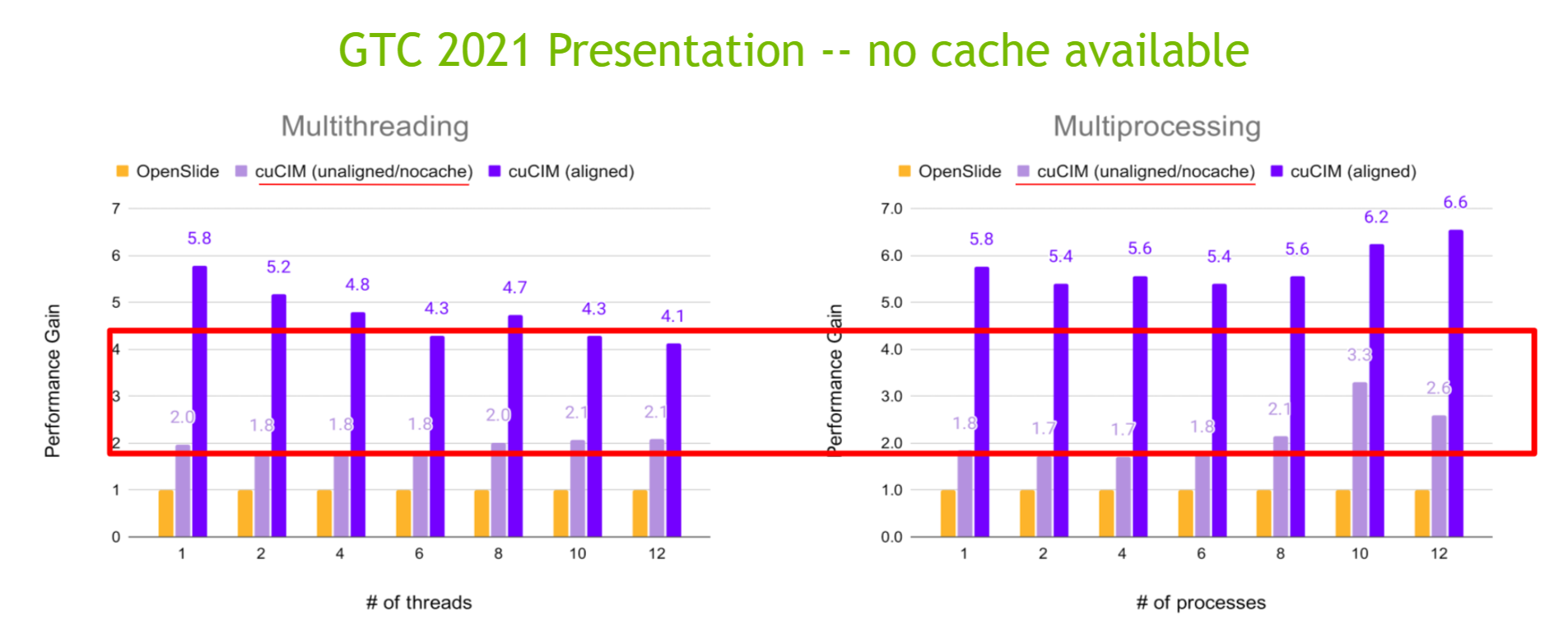
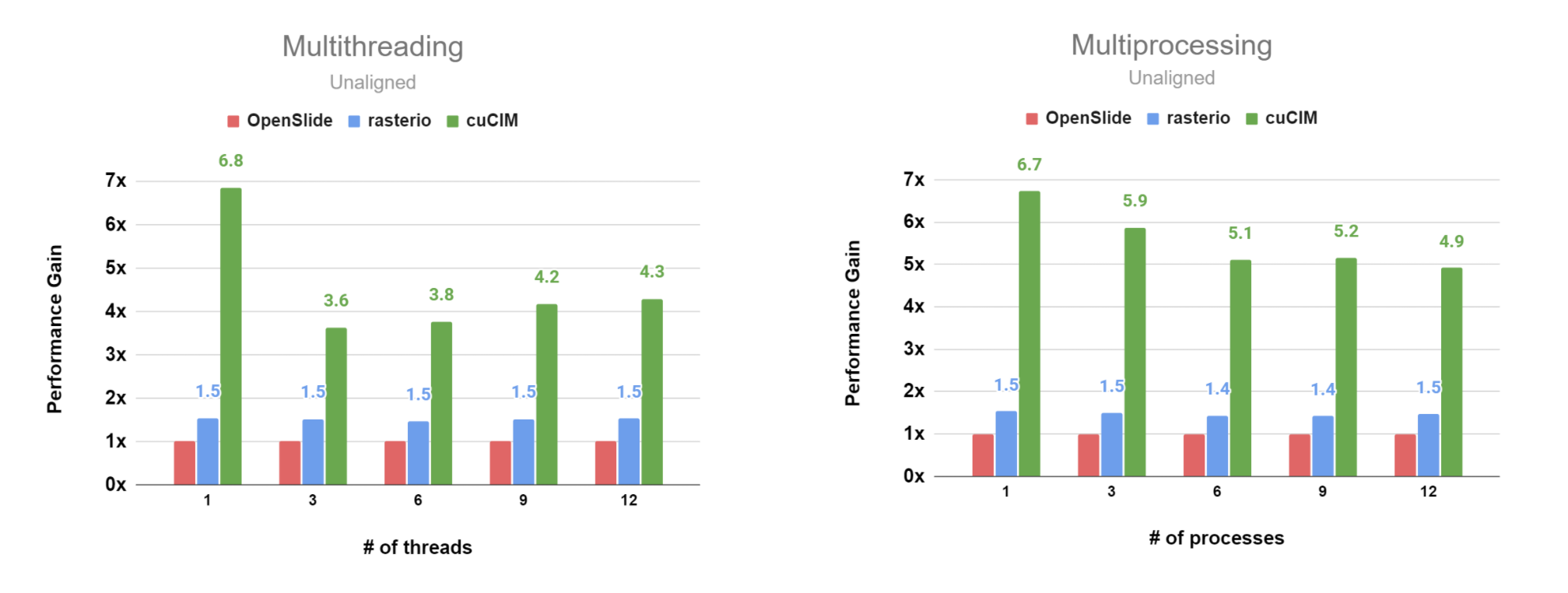
Unaligned Case (per_process, JPEG-compressed SVS file)
Unaligned Case (per_process, JPEG2000 RGB-compressed SVS file)
Unaligned Case (per_process, JPEG2000 YCbCr-compressed SVS file)
The detailed data is available here.
2. Add NVTX support for performance analysis
Important methods in cuCIM are instrumented with NVTX so can see performance bottlenecks easily with NSight systems.
Tracing can be enabled through config file or environment variable or through API and less than 1% performance overheads in normal execution.
Enabling Tracing
Through .cucim.json file
{
"profiler" : { "trace": true }
}Through Environment variable
CUCIM_TRACE=1 pythonThrough API
from cucim import CuImage
CuImage.profiler(trace=True)
#or
CuImage.profiler().trace(True)
CuImage.profiler().config
# {'trace': True}
CuImage.profiler().trace()
# True
CuImage.is_trace_enabled # this is simpler method.
# TrueProfiling with NVIDIA Nsight Systems
nsys profile -f true -t cuda,nvtx,osrt -s cpu -x true --trace-fork-before-exec true -o my_profile `which python` benchmark.py
# can add `--stats true`Then, execute nsight-sys to open the profile results (my_profile.qdrep).
With this feature, a bug in cuCIM v21.10.01 (thread contention in Cache) was found and fixed (#145).
3. Normalize operation
CUDA-based normalization operation is added. Normalization supports the following types.
- Simple range based normalization
- Arctangent based normalization
Arctangent-based normalization helps to stretch lower intensity pixels in the image slightly better than range-based normalization. If you look at its graph, there is a huge variation at certain lower intensities, but as intensities become higher, the curve becomes flatter. This helps in isolating regions like lungs (and regions within lungs) more efficiently. There can be separate use cases depending on the modality and the application.
Please check the test cases to see how you can use the operation.
🐛 Bug Fixes
- Load libcufile.so with RTLD_NODELETE flag (#177) @gigony
- Remove rmm/nvcc dependencies to fix cudaErrorUnsupportedPtxVersion error (#175) @gigony
- Do not compile code with nvcc if no CUDA kernel exists (#171) @gigony
- Fix a segmentation fault due to unloaded libcufile (#158) @gigony
- Fix thread contention in Cache (#145) @gigony
- Build with NumPy 1.17 (#148) @jakirkham
📖 Documentation
- Add Jupyter notebook for SVS Support (#147) @gigony
- Update change log for v21.10.01 (#142) @gigony
- update docs theme to pydata-sphinx-theme (#138) @quasiben
- Update Github links in README.md through script (#132) @gigony
- Fix GDS link in Jupyter notebook (#131) @gigony
- Update notebook for the interoperability with DALI (#127) @gigony
🛠️ Improvements
- Update
condarecipes for Enhanced Compatibility effort by (#164) @ajschmidt8 - Fix Changelog Merge Conflicts for
branch-21.12(#156) @ajschmidt8 - Add cucim.kit.cumed plugin with skeleton (#129) @gigony
- Update initial cpp unittests (#128) @gigony
- Optimize zoom out implementation with separate padding kernel (#125) @chirayuG-nvidia
- Do not force install linux-64 version of openslide-python (#124) @Ethyling
v21.10.00
Version 21.10.00 (Oct 06, 2021)
This version would be available through both Conda (https://anaconda.org/rapidsai/cucim) and PyPI package (https://pypi.org/project/cucim/21.10.0/).
🚀 New Features
- Add transforms for Digital Pathology (#100) @shekhardw @chirayuG-nvidia
- Enable GDS and Support Runtime Context (__enter__, __exit__) for CuFileDriver and CuImage (#106) @gigony
- Add a mechanism for user to know the availability of cucim.CuImage (#107) @gigony
- Support raw RGB tiled TIFF (#108) @gigony
1. Add high-performance transforms for Digital Pathology (#100)
We have implemented the following methods in this release.
cucim.core.operations.colormodule- color_jitter
cucim.core.operations.intensitymodule- rand_zoom
- scale_intensity_range
- zoom
cucim.core.operations.spatialmodule- image_flip
- image_rotate_90
- rand_image_flip
- rand_image_rotate_90
The above methods are exposed to MONAI (CuCIM Transforms #2932) through cucim.core.operations.expose.transform module
- color_jitter
- image_flip
- image_rotate_90
- scale_intensity_range
- zoom
- rand_zoom
- rand_image_flip
- rand_image_rotate_90
Special thanks to Chirayu Garg(@chirayuG-nvidia) for implementing C++ CUDA Kernels of the above transform methods!
2. Enable GDS and Support Runtime Context (__enter__, __exit__) for CuFileDriver and CuImage (#106)
Enabling GDS
GPU-Direct-Storage feature was disabled before (#3) because there was no public package available.
Now we re-enabled the feature. It's up to the user to install GDS libraries (GDS libraries are available through CUDA Toolkit >=11.x), or can install manually by following the instruction https://docs.nvidia.com/gpudirect-storage/troubleshooting-guide/index.html#gds-installing
If the GDS library is not available, the filesystem API still works with compatibility mode (our own implementation to do the same behavior).
Please see the Tutorial (Accessing File with GDS) for detail.
Support Runtime Context for CuFileDriver
CuFile Driver now can use Runtime Context so with statement is available.
with for cucim.clara.filesystem.open() would be available in the next release (v21.12.00)
from cucim.clara.filesystem import CuFileDriver
import cucim.clara.filesystem as fs
import os, cupy as cp, torch
# Assume a file ('nvme/input.raw') with size 10 in bytes
# Create a CuPy array with size 10 (in bytes)
cp_arr = cp.ones(10, dtype=cp.uint8)
# Create a PyTorch array with size 10 (in bytes)
cuda0 = torch.device('cuda:0')
torch_arr = torch.ones(10, dtype=torch.uint8, device=cuda0)
print(f"fs.is_gds_available(): {fs.is_gds_available()}")
# Using CuFileDriver
# (Opening a file with O_DIRECT flag is required for GDS)
fno = os.open("nvme/input.raw", os.O_RDONLY | os.O_DIRECT)
with CuFileDriver(fno) as fd:
# Read 8 bytes starting from file offset 0 into buffer offset 2
read_count = fd.pread(cp_arr, 8, 0, 2)
# Read 10 bytes starting from file offset 3
read_count = fd.pread(torch_arr, 10, 3)
# Another way of opening file with cuFile (>= v21.12.00)
# with fs.open("nvme/output.raw", "w") as fd:
# # Write 10 bytes from cp_array to file starting from offset 5
# write_count = fd.pwrite(cp_arr, 10, 5)
fd = fs.open("nvme/output.raw", "w")
# Write 10 bytes from cp_array to file starting from offset 5
write_count = fd.pwrite(cp_arr, 10, 5)
fd.close()Support Runtime Context for CuImage
CuImage class didn't have close() method (file handle is close when the object is destructed) but now close() method is available.
Once the file handle for the CuImage object is closed, it cannot call operations that require file access.
from cucim import CuImage
with CuImage("image.tif") as cuimg:
region = cuimg.read_region((0, 0), (100, 100))
print(region.shape)
region2 = cuimg.read_region((100, 100), (200, 200))[Plugin: cucim.kit.cuslide] Loading...
[Plugin: cucim.kit.cuslide] Loading the dynamic library from: /ssd/repo/cucim/python/cucim/src/cucim/clara/[email protected]
[Plugin: cucim.kit.cuslide] loaded successfully. Version: 0
Initializing plugin: cucim.kit.cuslide (interfaces: [cucim::io::IImageFormat v0.1]) (impl: cucim.kit.cuslide)
[100, 100, 3]
Traceback (most recent call last):
File "test_cucim.py", line 7, in <module>
region2 = cuimg.read_region((100, 100), (200, 200))
RuntimeError: [Error] The image file is closed!
[Plugin: cucim.kit.cuslide] Unloaded.3. Add a mechanism for users to know the availability of cucim.CuImage (#107)
Added cucim.is_available(module_name) method.
(module_name (str): Name of the module to check. (e.g. "skimage", "core", and "clara"))
Users can query if a specific module is available or not.
For example, if cucim.clara module which requires C shared library is not available:
>>> import cucim
>>> cucim.is_available()
False
>>> cucim.is_available("skimage")
True
>>> cucim.is_available("core")
True
>>> cucim.is_available("clara")
False
4. Support raw RGB tiled TIFF (#108)
Due to #17 ([BUG] Compression scheme 33003 tile decoding is not implemented), #19 (Check compression method used in the image) was merged so cuCIM has been handling only particular formats (only jpeg/deflate-compressed image).
If the input TIFF image has no-compressed RAW tile image, cuCIM showed the following error message:
RuntimeError: This format (compression: 1, sample_per_pixel: 1, planar_config: 1, photometric: 1) is not supported yet!.
https://github.com/Project-MONAI/MONAI/pull/2987/checks?check_run_id=3667530814#step:7:15641
Now cuCIM supports a raw RGB tiled TIFF with fast-path, re-allowing non-compressed image with slow-path (in case the input is not tiled RGB image).
This supports Project-MONAI/MONAI#2987.
🐛 Bug Fixes
📖 Documentation
- Forward-merge branch-21.08 to branch-21.10 (#88) @jakirkham
- Update PyPI cuCIM v21.08.01 README.md and CHANGELOG.md (#87) @gigony
🛠️ Improvements
- ENH Replace gpuci_conda_retry with gpuci_mamba_retry (#69) @dillon-cullinan
v21.08.01
Version 21.08.01 (August 10, 2021)
There are no new critical features in this release, but there are several enhancements/bug fixes.
We announced the cuCIM at the SciPy 2021 Conference (July 14). You can view the recorded video on YouTube (link).
🐛 Bug Fixes
- Remove int-type bug on Windows in skimage.measure.label (#72) @grlee77
- Fix missing array interface for associated_image() (#65) @gigony
- Handle zero-padding version string (#59) @gigony
- Remove invalid conda environment activation (#58) @ajschmidt8
📖 Documentation
🚀 New Features
- Pin
isorthook to 5.6.4 (#73) @charlesbluca - Add skimage.morphology.thin (#27) @grlee77
🛠️ Improvements
- Add SciPy 2021 to README (#79) @jakirkham
- Use more descriptive ElementwiseKernel names in cucim.skimage (#75) @grlee77
- Add initial Python unit/performance tests for TIFF loader module (#62) @gigony
- Fix
21.08forward-merge conflicts (#57) @ajschmidt8
v21.08.00
Version 21.08.00
There are no new critical features in this release, but there are several enhancements/bug fixes.
We announced the cuCIM at the SciPy 2021 Conference (July 14). You can view the recorded video on YouTube (link).
🐛 Bug Fixes
- Remove int-type bug on Windows in skimage.measure.label (#72) @grlee77
- Fix missing array interface for associated_image() (#65) @gigony
- Handle zero-padding version string (#59) @gigony
- Remove invalid conda environment activation (#58) @ajschmidt8
📖 Documentation
🚀 New Features
- Pin
isorthook to 5.6.4 (#73) @charlesbluca - Add skimage.morphology.thin (#27) @grlee77
🛠️ Improvements
- Add SciPy 2021 to README (#79) @jakirkham
- Use more descriptive ElementwiseKernel names in cucim.skimage (#75) @grlee77
- Add initial Python unit/performance tests for TIFF loader module (#62) @gigony
- Fix
21.08forward-merge conflicts (#57) @ajschmidt8
v21.06.00
Version 21.06.00 (June 8, 2021)
🚀 What's new?
- Change version scheme from SemVer to CalVer (#32)
- Implement cache mechanism (#48)
- Support
__cuda_array_interface__(#48)
Cache Mechanism
cuCIM now supports the cache mechanism for the unaligned use case.
Please check this notebook to see how to use the feature.
Support __cuda_array_interface__
In addition to the __array_interface__, cuCIM now supports __cuda_array_interface__ protocol.
Please check this notebook to see how to use the feature.
🐛 Fixes
- Check compression method used in the image for SVS format (#19)
- Avoid use of deprecated cp.float, cp.int, cp.bool (#29)
- Fix a memory leak in Deflate decoder (#48)
- Update
update-version.sh(#42) @ajschmidt8
🚀 Improvements
- Use CuPy 9 (#20)
- CuPy 9 is officially released so you don't need to install the release candidate (e.g., 9.0.0b3) anymore.
- Update environment variable used to determine
cuda_version(#43) @ajschmidt8 - Update version script to remove bump2version dependency (#41) @gigony
- Update changelog (#40) @ajschmidt8
- Update docs build script (#39) @ajschmidt8