-
Notifications
You must be signed in to change notification settings - Fork 125
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
- Loading branch information
Showing
1 changed file
with
390 additions
and
0 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,390 @@ | ||
| [](https://pypi.org/project/jarvis-tools/) | ||
| [](https://anaconda.org/conda-forge/jarvis-tools) | ||
| [](https://github.com/usnistgov/jarvis) | ||
| [](https://ci.appveyor.com/project/knc6/jarvis-63tl9) | ||
| [](https://github.com/usnistgov/jarvis) | ||
| [](https://github.com/usnistgov/jarvis) | ||
| [](https://codecov.io/gh/knc6/jarvis) | ||
| [](https://img.shields.io/pypi/dm/jarvis-tools.svg) | ||
| [](https://pepy.tech/badge/jarvis-tools) | ||
| [](https://doi.org/10.5281/zenodo.3903515) | ||
| [](https://github.com/usnistgov/jarvis) | ||
| [](https://github.com/usnistgov/jarvis) | ||
| [](https://figshare.com/authors/Kamal_Choudhary/4445539) | ||
| [](https://jarvis-tools.readthedocs.io) | ||
| [](https://github.com/JARVIS-Materials-Design/jarvis-tools-notebooks) | ||
| <!-- [](https://travis-ci.org/usnistgov/jarvis) --> | ||
| ------------------------------------------------------------------------ | ||
|
|
||
| # Table of Contents | ||
| * [Introduction](#intro) | ||
| * [Documentation](#doc) | ||
| * [Capabilities](#cap) | ||
| * [Installation](#install) | ||
| * [Example function](#example) | ||
| * [Citation](#cite) | ||
| * [References](#refs) | ||
| * [How to contribute](#contrib) | ||
| * [Correspondence](#corres) | ||
| * [Funding support](#fund) | ||
| * [Code of conduct](#conduct) | ||
| * [Module structure](#module) | ||
|
|
||
| <a name="intro"></a> | ||
| # JARVIS-Tools (Introduction) | ||
|
|
||
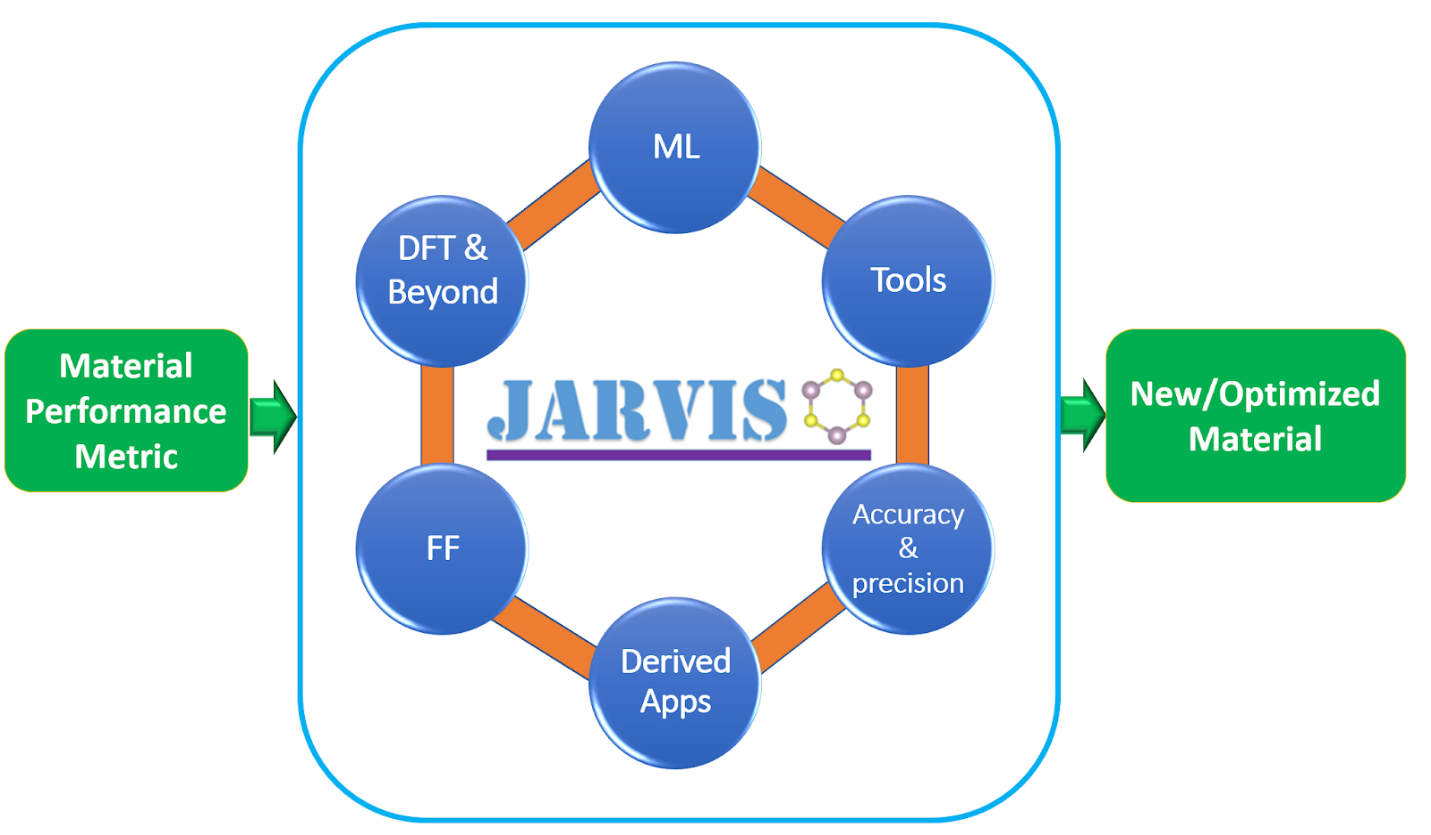
| The JARVIS-Tools is an open-access software package for atomistic | ||
| data-driven materials design. JARVIS-Tools can be used for a) setting up | ||
| calculations, b) analysis and informatics, c) plotting, d) database | ||
| development and e) web-page development. | ||
|
|
||
| JARVIS-Tools empowers NIST-JARVIS (Joint Automated Repository for | ||
| Various Integrated Simulations) repository which is an integrated | ||
| framework for computational science using density functional theory, | ||
| classical force-field/molecular dynamics and machine-learning. The | ||
| NIST-JARVIS official website is: <https://jarvis.nist.gov> . This | ||
| project is a part of the Materials Genome Initiative (MGI) at NIST | ||
| (<https://mgi.nist.gov/>). | ||
|
|
||
| For more details, checkout our latest article: [The joint automated | ||
| repository for various integrated simulations (JARVIS) for data-driven | ||
| materials design](https://www.nature.com/articles/s41524-020-00440-1) | ||
| and [YouTube | ||
| videos](https://www.youtube.com/watch?v=P0ZcHXOC6W0&feature=emb_title&ab_channel=JARVIS-repository) | ||
|
|
||
| [](https://jarvis.nist.gov/) | ||
|
|
||
| <a name="doc"></a> | ||
| ## Documentation | ||
|
|
||
| > <https://jarvis-tools.readthedocs.io> | ||
| <a name="cap"></a> | ||
| ## Capabilities | ||
|
|
||
| - **Software workflow tasks for preprcessing, executing and | ||
| post-processing**: VASP, Quantum Espresso, Wien2k BoltzTrap, | ||
| Wannier90, LAMMPS, Scikit-learn, TensorFlow, LightGBM, Qiskit, | ||
| Tequila, Pennylane, DGL, PyTorch. | ||
| - **Several examples**: Notebooks and test scripts to explain the | ||
| package. | ||
| - **Several analysis tools**: Atomic structure, Electronic structure, | ||
| Spacegroup, Diffraction, 2D materials and other vdW bonded systems, | ||
| Mechanical, Optoelectronic, Topological, Solar-cell, Thermoelectric, | ||
| Piezoelectric, Dielectric, STM, Phonon, Dark matter, Wannier tight | ||
| binding models, Point defects, Heterostructures, Magnetic ordering, | ||
| Images, Spectrum etc. | ||
| - **Database upload and download**: Download JARVIS databases such as | ||
| JARVIS-DFT, FF, ML, WannierTB, Solar, STM and also external | ||
| databases such as Materials project, OQMD, AFLOW etc. | ||
| - **Access raw input/output files**: Download input/ouput files for | ||
| JARVIS-databases to enhance reproducibility. | ||
| - **Train machine learning models**: Use different descriptors, graphs | ||
| and datasets for training machine learning models. | ||
| - **HPC clusters**: Torque/PBS and SLURM. | ||
| - **Available datasets**: [Summary of several | ||
| datasets](https://github.com/usnistgov/jarvis/blob/master/DatasetSummary.rst) | ||
| . | ||
| <a name="install"></a> | ||
| ## Installation | ||
|
|
||
| - We recommend installing miniconda environment from | ||
| <https://conda.io/miniconda.html> : | ||
|
|
||
| bash Miniconda3-latest-Linux-x86_64.sh (for linux) | ||
| bash Miniconda3-latest-MacOSX-x86_64.sh (for Mac) | ||
| Download 32/64 bit python 3.8 miniconda exe and install (for windows) | ||
| Now, let's make a conda environment just for JARVIS:: | ||
| conda create --name my_jarvis python=3.8 | ||
| source activate my_jarvis | ||
|
|
||
| - Method-1: Installation using pip: | ||
|
|
||
| pip install -U jarvis-tools | ||
|
|
||
| - Method-2: Installation using conda: | ||
|
|
||
| conda install -c conda-forge jarvis-tools | ||
|
|
||
| - Method-3: Installation using setup.py: | ||
|
|
||
| pip install numpy scipy matplotlib | ||
| git clone https://github.com/usnistgov/jarvis.git | ||
| cd jarvis | ||
| python setup.py install | ||
|
|
||
| - Note on installing additional dependencies for all modules to | ||
| function: | ||
|
|
||
| pip install -r dev-requirements.txt | ||
|
|
||
| <a name="example"></a> | ||
| ## Example function | ||
| ``` | ||
| from jarvis.core.atoms import Atoms | ||
| box = [[2.715, 2.715, 0], [0, 2.715, 2.715], [2.715, 0, 2.715]] | ||
| coords = [[0, 0, 0], [0.25, 0.25, 0.25]] | ||
| elements = ["Si", "Si"] | ||
| Si = Atoms(lattice_mat=box, coords=coords, elements=elements) | ||
| density = round(Si.density,2) | ||
| print (density) | ||
| 2.33 | ||
| from jarvis.db.figshare import data | ||
| dft_3d = data(dataset='dft_3d') | ||
| print (len(dft_3d)) | ||
| 75993 | ||
| from jarvis.io.vasp.inputs import Poscar | ||
| for i in dft_3d: | ||
| atoms = Atoms.from_dict(i['atoms']) | ||
| poscar = Poscar(atoms) | ||
| jid = i['jid'] | ||
| filename = 'POSCAR-'+jid+'.vasp' | ||
| poscar.write_file(filename) | ||
| dft_2d = data(dataset='dft_2d') | ||
| print (len(dft_2d)) | ||
| 1109 | ||
| for i in dft_2d: | ||
| atoms = Atoms.from_dict(i['atoms']) | ||
| poscar = Poscar(atoms) | ||
| jid = i['jid'] | ||
| filename = 'POSCAR-'+jid+'.vasp' | ||
| poscar.write_file(filename) | ||
| # Example to parse DOS data from JARVIS-DFT webpages | ||
| from jarvis.db.webpages import Webpage | ||
| from jarvis.core.spectrum import Spectrum | ||
| import numpy as np | ||
| new_dist=np.arange(-5, 10, 0.05) | ||
| all_atoms = [] | ||
| all_dos_up = [] | ||
| all_jids = [] | ||
| for ii,i in enumerate(dft_3d): | ||
| all_jids.append(i['jid']) | ||
| try: | ||
| w = Webpage(jid=i['jid']) | ||
| edos_data = w.get_dft_electron_dos() | ||
| ens = np.array(edos_data['edos_energies'].strip("'").split(','),dtype='float') | ||
| tot_dos_up = np.array(edos_data['total_edos_up'].strip("'").split(','),dtype='float') | ||
| s = Spectrum(x=ens,y=tot_dos_up) | ||
| interp = s.get_interpolated_values(new_dist=new_dist) | ||
| atoms=Atoms.from_dict(i['atoms']) | ||
| ase_atoms=atoms.ase_converter() | ||
| all_dos_up.append(interp) | ||
| all_atoms.append(atoms) | ||
| all_jids.append(i['jid']) | ||
| filename=i['jid']+'.cif' | ||
| atoms.write_cif(filename) | ||
| break | ||
| except Exception as exp : | ||
| print (exp,i['jid']) | ||
| pass | ||
| ``` | ||
|
|
||
| Find more examples at | ||
|
|
||
| > 1. <https://jarvis-tools.readthedocs.io/en/master/tutorials.html> | ||
| > 2. <https://github.com/JARVIS-Materials-Design/jarvis-tools-notebooks> | ||
| > 3. <https://github.com/usnistgov/jarvis/tree/master/jarvis/tests/testfiles> | ||
| <a name="cite"></a> | ||
| ## Citing | ||
|
|
||
| Please cite the following if you happen to use JARVIS-Tools for a | ||
| publication. | ||
|
|
||
| <https://www.nature.com/articles/s41524-020-00440-1> | ||
|
|
||
| > Choudhary, K. et al. The joint automated repository for various | ||
| > integrated simulations (JARVIS) for data-driven materials design. npj | ||
| > Computational Materials, 6(1), 1-13 (2020). | ||
| <a name="refs"></a> | ||
| ## References | ||
|
|
||
| Please see [Publications related to | ||
| JARVIS-Tools](https://jarvis-tools.readthedocs.io/en/master/publications.html) | ||
|
|
||
| <a name="contrib"></a> | ||
| ## How to contribute | ||
|
|
||
| [](http://makeapullrequest.com) | ||
|
|
||
| For detailed instructions, please see [Contribution | ||
| instructions](https://github.com/usnistgov/jarvis/blob/master/Contribution.rst) | ||
|
|
||
| <a name="corres"></a> | ||
| ## Correspondence | ||
|
|
||
| Please report bugs as Github issues | ||
| (<https://github.com/usnistgov/jarvis/issues>) or email to | ||
| <[email protected]>. | ||
|
|
||
| <a name="fund"></a> | ||
| ## Funding support | ||
|
|
||
| NIST-MGI (<https://www.nist.gov/mgi>). | ||
|
|
||
| <a name="conduct"></a> | ||
| ## Code of conduct | ||
|
|
||
| Please see [Code of | ||
| conduct](https://github.com/usnistgov/jarvis/blob/master/CODE_OF_CONDUCT.md) | ||
|
|
||
| <a name="module"></a> | ||
| ## Module structure | ||
|
|
||
| jarvis/ | ||
| ├── ai | ||
| │ ├── descriptors | ||
| │ │ ├── cfid.py | ||
| │ │ ├── coulomb.py | ||
| │ ├── gcn | ||
| │ ├── pkgs | ||
| │ │ ├── lgbm | ||
| │ │ │ ├── classification.py | ||
| │ │ │ └── regression.py | ||
| │ │ ├── sklearn | ||
| │ │ │ ├── classification.py | ||
| │ │ │ ├── hyper_params.py | ||
| │ │ │ └── regression.py | ||
| │ │ └── utils.py | ||
| │ ├── uncertainty | ||
| │ │ └── lgbm_quantile_uncertainty.py | ||
| ├── analysis | ||
| │ ├── darkmatter | ||
| │ │ └── metrics.py | ||
| │ ├── defects | ||
| │ │ ├── surface.py | ||
| │ │ └── vacancy.py | ||
| │ ├── diffraction | ||
| │ │ └── xrd.py | ||
| │ ├── elastic | ||
| │ │ └── tensor.py | ||
| │ ├── interface | ||
| │ │ └── zur.py | ||
| │ ├── magnetism | ||
| │ │ └── magmom_setup.py | ||
| │ ├── periodic | ||
| │ │ └── ptable.py | ||
| │ ├── phonon | ||
| │ │ ├── force_constants.py | ||
| │ │ └── ir.py | ||
| │ ├── solarefficiency | ||
| │ │ └── solar.py | ||
| │ ├── stm | ||
| │ │ └── tersoff_hamann.py | ||
| │ ├── structure | ||
| │ │ ├── neighbors.py | ||
| │ │ ├── spacegroup.py | ||
| │ ├── thermodynamics | ||
| │ │ ├── energetics.py | ||
| │ ├── topological | ||
| │ │ └── spillage.py | ||
| ├── core | ||
| │ ├── atoms.py | ||
| │ ├── composition.py | ||
| │ ├── graphs.py | ||
| │ ├── image.py | ||
| │ ├── kpoints.py | ||
| │ ├── lattice.py | ||
| │ ├── pdb_atoms.py | ||
| │ ├── specie.py | ||
| │ ├── spectrum.py | ||
| │ └── utils.py | ||
| ├── db | ||
| │ ├── figshare.py | ||
| │ ├── jsonutils.py | ||
| │ ├── lammps_to_xml.py | ||
| │ ├── restapi.py | ||
| │ ├── vasp_to_xml.py | ||
| │ └── webpages.py | ||
| ├── examples | ||
| │ ├── lammps | ||
| │ │ ├── jff_test.py | ||
| │ │ ├── Al03.eam.alloy_nist.tgz | ||
| │ ├── vasp | ||
| │ │ ├── dft_test.py | ||
| │ │ ├── SiOptb88.tgz | ||
| ├── io | ||
| │ ├── boltztrap | ||
| │ │ ├── inputs.py | ||
| │ │ └── outputs.py | ||
| │ ├── calphad | ||
| │ │ └── write_decorated_poscar.py | ||
| │ ├── lammps | ||
| │ │ ├── inputs.py | ||
| │ │ └── outputs.py | ||
| │ ├── pennylane | ||
| │ │ ├── inputs.py | ||
| │ ├── phonopy | ||
| │ │ ├── fcmat2hr.py | ||
| │ │ ├── inputs.py | ||
| │ │ └── outputs.py | ||
| │ ├── qe | ||
| │ │ ├── inputs.py | ||
| │ │ └── outputs.py | ||
| │ ├── qiskit | ||
| │ │ ├── inputs.py | ||
| │ ├── tequile | ||
| │ │ ├── inputs.py | ||
| │ ├── vasp | ||
| │ │ ├── inputs.py | ||
| │ │ └── outputs.py | ||
| │ ├── wannier | ||
| │ │ ├── inputs.py | ||
| │ │ └── outputs.py | ||
| │ ├── wanniertools | ||
| │ │ ├── inputs.py | ||
| │ │ └── outputs.py | ||
| │ ├── wien2k | ||
| │ │ ├── inputs.py | ||
| │ │ ├── outputs.py | ||
| ├── tasks | ||
| │ ├── boltztrap | ||
| │ │ └── run.py | ||
| │ ├── lammps | ||
| │ │ ├── templates | ||
| │ │ └── lammps.py | ||
| │ ├── phonopy | ||
| │ │ └── run.py | ||
| │ ├── vasp | ||
| │ │ └── vasp.py | ||
| │ ├── queue_jobs.py | ||
| ├── tests | ||
| │ ├── testfiles | ||
| │ │ ├── ai | ||
| │ │ ├── analysis | ||
| │ │ │ ├── darkmatter | ||
| │ │ │ ├── defects | ||
| │ │ │ ├── elastic | ||
| │ │ │ ├── interface | ||
| │ │ │ ├── magnetism | ||
| │ │ │ ├── periodic | ||
| │ │ │ ├── phonon | ||
| │ │ │ ├── solar | ||
| │ │ │ ├── stm | ||
| │ │ │ ├── structure | ||
| │ │ │ ├── thermodynamics | ||
| │ │ │ ├── topological | ||
| │ │ ├── core | ||
| │ │ ├── db | ||
| │ │ ├── io | ||
| │ │ │ ├── boltztrap | ||
| │ │ │ ├── calphad | ||
| │ │ │ ├── lammps | ||
| │ │ │ ├── pennylane | ||
| │ │ │ ├── phonopy | ||
| │ │ │ ├── qiskit | ||
| │ │ │ ├── qe | ||
| │ │ │ ├── tequila | ||
| │ │ │ ├── vasp | ||
| │ │ │ ├── wannier | ||
| │ │ │ ├── wanniertools | ||
| │ │ │ ├── wien2k | ||
| │ │ ├── tasks | ||
| │ │ │ ├── test_lammps.py | ||
| │ │ │ └── test_vasp.py | ||
| └── README.rst |